Mi SciELO
Servicios Personalizados
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista de Salud Animal
versión On-line ISSN 2224-4700
Rev Salud Anim. vol.38 no.2 La Habana mayo-ago. 2016
ORIGINAL ARTICLE
Prevalence of Mollicutes in Cell Cultures: experience in Cuba
Prevalencia de Mollicutes en cultivos celulares: experiencia en Cuba
Evelyn Lobo RiveroI*, Anisleidy Pérez CastilloI, Arianna Duque OrtizI, Yaima Burgher PulgarónI, Lucas Miranda MarquesII, Jorge TimenetskyII
IMYCOLAB, Laboratorio para el diagnóstico de micoplasmas, Departamento de Microbiología, Centro Nacional de Sanidad Agropecuaria (CENSA), Carretera de Tapaste y Autopista Nacional, San José de las Lajas, Mayabeque, Cuba.
IIDepartamento de Microbiología, Instituto de Ciências Biomédicas, Universidade de São Paulo, Brazil.
ABSTRACT
Different mollicutes species of the Mycoplasma and Acholeplasma genera can contaminate the cell cultures and raw materials commonly used in the manufacture of a variety of biological and therapeutical products. In this study, the presence of mollicutes was detected in 19 of 50 cell culture samples analyzed using PCR and microbiological cultures. The species most frequently detected was M. fermentans (31.5%), followed by A. laidlawii (26.3%), M. orale (15.7%), M. hyorhinis (15.7%), and M. salivarium (10, 5%). A. laidlawii was present as the infecting agent only in 10.5% of the samples. These results showed the main species of contaminants, which suggested mollicutes multiple origin in the source of infection.
Key words: mollicutes, Mycoplasmas, Acholeplasma, cell cultures, diagnostic.
RESUMEN
Diferentes especies de mollicutes, de los géneros Mycoplasma y Acholeplasma, pueden contaminar los cultivos celulares y las materias primas que se utilizan comúnmente en la fabricación de una variedad de productos biológicos y terapéuticos. En el presente estudio se detectó la presencia de mollicutes en 19 de 50 muestras analizadas de cultivo celular a través de PCR y cultivo microbiológico. Las especies detectadas con más frecuencia fueron M. fermentans (31,5%), seguido por A. laidlawii (26,3%), M. orale (15,7%), M. hyorhinis (15,7%) y M. salivarium (10,5%). A. laidlawii estaba presente como el agente infectante solo en 10,5% de las muestras. Estos resultados muestran las principales especies de mollicutes contaminantes, lo que sugiere un origen múltiple en la fuente de infección.
Palabras clave: mollicutes, Mycoplasmas, Acholeplasmas, cultivos celulares, diagnóstico.
INTRODUCTION
Mycoplasmas (Mollicutes) remain the smallest free-living bacteria found in animals including humans, plants and insects (1). Mycoplasma and Acholeplasma are the main genera that contaminate cell cultures and their substrates, as well as the raw materials commonly used for manufacturing a variety of biological and therapeutic products (2). Mycoplasma contamination, even at low load, at the initial steps of manufacture, generally results in the loss of the whole batch of the product due to poor cell growth or regulatory safety concerns.
The first isolation of mycoplasma from a cell culture was reported in 1956 (3). At present, about 20 species have been identified (4). Acholeplasma laidlawii, Mycoplasma arginini, M.orale, M. salivarium, M.fermentans, and M. hyorhins have been detected in about 95% of cell cultures. Species such as Ureaplasma urealyticum, M. pneumoniae and M. pirum are rarely present in cell cultures and some of them have been isolated only once (1, 5).
In most cases, mycoplasma infection originates from contaminated animal serum, but contaminated aerosols produced at the laboratory helps their spread. The frequency of mycoplasmas in cell cultures depends on sampling, institution and the time for testing (6). Mycoplasma contamination cause many cytogenetic effects in cell cultures, leading to unreliable experimental results and potential harmful biological products (7). Therefore, the result for mycoplasma contamination testing must be strongly confident for the quality of biotechnological products (8). In addition, as consequence of a wrong result, there are economic and labor time losses.
In Cuba, the biotechnology industry is increasingly being developed. Therefore, quality standards for detection of mollicutes as contaminants of biotech products have been established for biomedical application. However, the identification of mollicutes species in cell cultures must also be added to the regular testing procedures. This approach will help to a better control of the origin of such contaminations (2). The aim of this study was to explore the status of mycoplasma contaminations in cell cultures sent to our laboratory to be tested.
MATERIAL AND METHODS
Samples
The following mycoplasma ATCC strains were used as controls: U. urealyticum T960, A. laidlawii P8, M. pneumoniae-FH, M.genitalium G37, M. hominis PG-21, M.hyorhinis BTS7, M. orale CH19299, M.salivarium PG-20, M. buccale CH202247, M. fermentans PG-18, M. pulmonis PG-34, and M. arginini G230.
Sample
Fifty cell culture samples were tested in 2014-2015; some cell cultures did not show morphological alterations and were monitored for the first time. The samples were from five laboratories in Havana, Cuba,
Culture
Each cell sample was inoculated in liquid and solid Hyflick´s medium. The cultures were incubated for 15 days at 37ºC under aerobic and anaerobic conditions (2). The microorganism was presumptively identified based on pH shifts of the broth without turbidity, production of «fried egg» colonies on Hyflick´s medium agar plates and positive subculture after filtration of the initial culture through 0.22-mm membranes (9).
Polymerase chain reaction
The technique used was that described by Timenetsky et al. (6). The targeted DNA was extracted from the cell culture samples and from cultures of the reference strains by boiling 1 ml of each sample.
Twenty-five pmoles of each primer (Table 1), 1U Taq DNA polymerase (Biotools), 1.4; 1.8 and 1.6 mM MgCl2 (Promega) for M. salivarium, M. arginini and M. hyorhinis, respectively, 200 mM of each dNTP (Promega), 1 ml DNA extracted from the cell culture sample and ultrapure water to a final volume of 50 mL. The amplification was made in a thermocycler (Eppendorf). It was programmed for 40 cycles at 94ºC for 30 s, 55ºC for 30 s, and 72ºC for 60 s and a final step at 72ºC for 5 min.For the detection of M. fermentans, M.orale and A. laidlawii, the reaction mixture contained 40 pmol of each primer, 1 U Taq DNA polymerase (Biotools), 1.2 mM MgCl2 (Promega), 200 mM of each dNTP (Promega), 1 mL DNA extracted from the cell culture sample, and ultrapure water to a final volume of 50 mL. The amplification was made in a thermocycler (Eppendorf). It was programmed for one cycle at 95ºC for 15 min, 30 cycles at 95ºC for 30 s, 64ºC for 90 s, and 72ºC for 90 s, and a final step at 72ºC for 10 min.
A positive control (DNA of each reference strains) and a negative control (ultrapure water) were added to all amplifications.
RESULTS AND DISCUSSION
Mycoplasma-infected cell lines are themselves the single most important source for further spreading of contamination. This is due to the high concentration of mycoplasmas in infected cultures, and the prolonged survival of dried mycoplasmas (5). Operator-induced contamination is also a potential issue. Mycoplasmas spread by using laboratory equipment, media, or reagents that have been contaminated (8).
In the present study, 19 (38%) cell culture samples out of 50 were positive for mycoplasma culturing; 15 samples (78.9%) presented pH shifts in the broths at 48 hours, no pH shifts were observed in the remaining samples. However, when the samples were subcultured from the broths to the agar plates, all 19 produced «fried egg» colonies. Interestingly, 75.8% of the infected cell cultures presented at least two mycoplasma species, and 15.78% of the samples were infected with three species according to the biochemical test results.
According to Neto et al. (8), up to 87% of the cellular cultures could be contaminated by mycoplasmas. The contamination percentage variation found in the literature was related to the size of the sample population studied, contamination control practices, and efficiency of the detection assays used; in our case, the percentage was in agreement with international reports.
The mollicutes species detected in the present study derived from animals, humans, or both, and the diversity of mycoplasma species in the same cell culture indicated the occurrence of different initial infection sources. In our case, we agree with Kazemiha et al. (4) about the subculturing of a cell culture among laboratories over time, which, due to successive sharing, may explain the detection of multiple mycoplasma species. Mycoplasma diversity accumulates over time mainly due to failure to control the infection.
On the other hand, because many infectious agents are not easily cultivable, PCR has been shown to be an efficient methodology for detecting biological contaminants in cell culture and its supplies (10). Moreover, the PCR technique has attracted much attention in the detection of cell culture contaminants because it is fast, robust, highly sensitive and specific compared with traditional techniques.
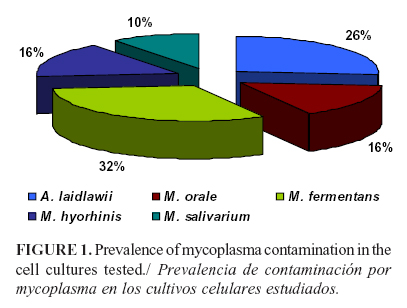
The results by PCR showed that the most frequent species was M. fermentans in 31.5% samples, followed by A. laidlawii in 26.3%, M. orale in 15.7%, M. hyorhinis in 15.7%, and M. salivarium in 10.5%. A. laidlawii was detected as the single mollicutes in 10.5% samples (Figure 1).
In fact, M. fermentans and A. laidlawii were also identified by Uphoff and Drexler (5). For A. laidlawii, the result was similar to that mentioned by Timenetsky et al. (6), and in the case of M. orale, the percentage obtained in our study was similar to that reported by Kazemiha et al. (4), who indicated values of 12.5%. Moreover, M. hyorhinis percentage in our case was 15.7, which was different from the percentages between 42 and 32 reported by other authors (11, 12). In this case, this percentage differences could be explained by the type of sample worked because previous studies reported trypsin and no cell cultures as the major source of contamination of M. hyorhinis (9).
In most cases, the contamination was through mycoplasmas derived from animal sera, mainly from contaminated cattle, as well as from aerosols derived from humans due to non-aseptic practices in the laboratory environments (13), justifying why such inputs were important to choose.
Technical procedures in a laboratory are the main sources of M. orale, M. fermentans, and M. hominis (3). These mycoplasmas account for more than half of all mycoplasma infections in cell cultures, and physiologically they are found in the human oropharyngeal tract (11). M. arginini and A. laidlawii are species originated from fetal bovine serum (FBS) or newborn bovine serum (NBS). Although M hyorhynis has a swine origin, mycoplasmas have never been isolated from this solution and their DNA has been rarely detected. Trypsin has mycoplasmicide activity (9).
There is a number of different sources for mycoplasma contamination in cell cultures associated with human, bovine, and swine species (14). M. fermentans was considered a normal inhabitant of the human urogenital tract and it is a fastidious species, a fact that impaired its isolation in the past. In 1986, M. fermentans was considered as a co-factor for the development of AIDS in HIV-positive individuals, a fact that, in turn, attracted the interest of the scientific community (15). Subsequently, this microorganism has been detected in or associated most frequently with tissues and blood of individuals with diseases poorly studied or with unknown etiology (14). In our case, the increase in the frequency of M. fermentans in cell cultures can be explained by the increasing use of human blood cells or tissues for primary culture.
Regarding A. laidlawii, it is a commensal of the mucosa in the upper respiratory and urogenital tract of many animal and bird species. Its potential pathogenicity has been the subject of investigation particularly with regard to bovine mastitis and spontaneous abortion in farm animals (9). Generally, it is regarded as non-pathogenic but may cause opportunistic, often transient, infections; the only reported incidence of human isolation was from an infected burn wound (14). The high incidence of A. laidlawii in cell cultures in this work seems,,to be in direct correlation with the use of fetal or newborn bovine serum.
Potential undetected contamination of these products or process intermediates with mycoplasmas presents a potential safety risk for patients and a business risk for producers of biopharmaceuticals (1). To minimize these risks, monitoring for adventitious agents, such as mycoplasmas, is performed during the manufacture of biologics produced in cell culture substrates.
The results of this study show the importance of mollicutes diagnosis in cell cultures, as they remain one of the most common contaminations. In addition, it is also important to know the possible sources of infection to help control and taking action to prevent the spread to other cell lines and biological products.
ACKNOWLEDGMENTS
Special thanks are due to CAPES-Brasil and to Dr. Jorge Timenetsky from the University of São Paulo in the make this work.
REFERENCES
1. Volokhov DV, Laurie J, Graham KA, Brorson E, Chizhikov V. Mycoplasma testing of cell substrates and biologics: Review of alternative non-microbiological techniques. Mol Cell Probe. 2011;25:69-77.
2. Lozada Y, Betancourt A, Lobo E. Detección confiable de micoplasmas como contaminantes en cultivos biológicos. Tesis en opción al grado de Master en Microbiología veterinaria (MSc), Mayabeque, Cuba. 2013.
3. Robinson LB, Wichelhausen RH. Contamination of human cell cultures by pleuropneumonialike organisms. Science. 1956;124:1147-1148.
4. Kazemiha VM, Shokrgozar MA, Arabestani MR, Moghadam MS, Azari S, Shokri F. PCR-based detection and eradication of mycoplasmal infections from various mammalian cell lines: a local experience. Cytotechnology. 2009;61:117-124. DOI 10.1007/s10616-010-9252-6.
5. Uphoff CC, Drexler HG. Comparative PCR analysis for detection of mycoplasma infections in continuous cell lines. Cancer Cell Culture: Methods and Protocols, 2nd Edition, Methods in Molecular Biology. 2006:731.
6. Timenetsky J, Santos LM, Buzinhani M, Mettifogo E. Detection of multiple mycoplasma infection in cell cultures by PCR. Brazilian Journal of Medical and Biological Research. 2006;39:907-914.
7. Schmitt M, Pawlita M. High-throughput detection and multiplex identification of cell contaminations. Nucleic Acids Res. 2004;37:119-126.
8. Netto C, Thomaz V, Sepulveda L, Oliveira GH, Timenetsky J. Quality Control of Biotechnological Inputs Detecting Mycoplasma. Braz Arch Biol Technol. 2015;58(2):239-243.
9. Windsor HM, Windsor GD, Noordergraaf JH. The growth and long-term survival of Acholeplasma laidlawii in media products used in biopharmaceutical manufacturing. Biologicals. 2010;38:204-210.
10.Van Kuppeveld FJM, Johansson KE, Galama JMD, Kissing J, Bölske G, et al. Detection of Mycoplasma contamination in cell cultures by mycoplasma group-specific PCR. Appl Environ Microbiol. 1994;60:149-152.
11.Camargos MF, Oliveira AM, Fonseca Junior AA, Rivetti AV, Motta PMC, Assis RA. Aplicação da reação em cadeia da polimerase para detecção de Mycoplasma spp na rotina de cultivos celulares. Ciênc Anim Bras. 2008;9:786-790.
12.Rivera-Tapia JA, Muñoz GZ, Román C. Contaminación por micoplasmas en cultivos celulares. Anales Médicos. 2006;51(3):109-112.
13.Rottem S. Interaction of mycoplasmas with host cells. Physiol Rev. 2003;83:413-432.
14.Pinheiro de Oliveira TF, Fonseca AJr, Fernandes M, Camargos M, Macedo de Oliveira A, Pinto AC, et al. Detection of contaminants in cell cultures, sera and trypsin. Biologicals. 2013;41:407-414.
15.Shu H-W, Liu T-T, Chan H-I, Liu Y-M, Wu K-M, et al. Complexity of the Mycoplasma fermentans M64 Genome and Metabolic Essentiality and Diversity among Mycoplasmas. PLoS ONE. 2012;7(4): e32940. doi:10.1371/journal.pone.0032940.
Recibido: 4-11-2015.
Aceptado: 9-5-2016.
* Corresponding author: Evelyn Lobo Rivero. E-mail: elobo@censa.edu.cu












 Curriculum ScienTI
Curriculum ScienTI