Mi SciELO
Servicios Personalizados
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Biotecnología Aplicada
versión On-line ISSN 1027-2852
Biotecnol Apl vol.29 no.3 La Habana jul.-set. 2012
RESEARCH
Effect of human epidermal growth factor on the tumor cell line A431: in vivo analysis of tumor growth inhibition and gene expression
Efecto del factor de crecimiento epidérmico humano en la línea celular tumoral A431: análisis de la inhibición del crecimiento tumoral y la expresión génica in vivo
Isabel A Guillén1, Hanlet Camacho1, Maria E Fernández1, Daniel O Palenzuela1, Lincidio Pérez2, Maria E Ochagavia3, Julio A Ancizar4, Angela D Tuero5, Laritza Gorovaya4, Osmani Mendoza4, Tamara Díaz1, Julio R Fernández1, Juan Roca1, Karelia Cosme4, Gerardo E Guillén-Nieto6, Luis Herrera7, Jorge Berlanga8, Lidia I Novoa1
1 Departamento de Farmacogenómica, Dirección de Investigaciones Biomédicas. Centro de Ingeniería Genética y Biotecnología, CIGB. Ave. 31 e/ 158 y 190, Playa, CP 10 600, La Habana, Cuba.
2 Departamento de Cáncer, Dirección de Investigaciones Biomédicas, Centro de Ingeniería Genética y Biotecnología. La Habana, Cuba.
3 Departamento de Bioinformática, Dirección de Investigaciones Biomédicas. Centro de Ingeniería Genética y Biotecnología. La Habana, Cuba.
4 Bioterio. Centro de Ingeniería Genética y Biotecnología. La Habana, Cuba.
5 Departamento de Estadísticas, Dirección de Ensayos Clínicos. Centro de Ingeniería Genética y Biotecnología. La Habana, Cuba.
6 Departamento de Vacunas, Dirección de Investigaciones Biomédicas. Centro de Ingeniería Genética y Biotecnología. La Habana, Cuba.
7 Dirección General. Centro de Ingeniería Genética y Biotecnología. La Habana, Cuba.
8 Departamento de Cicatrización y Citoprotección. Dirección de Investigaciones Biomédicas. Centro de Ingeniería Genética y Biotecnología. La Habana, Cuba.
ABSTRACT
The historical tag of the epidermal growth factor (EGF) as carcinogenesis promoter has not been uniformly reproduced, since, in some experiments, malignant cells are bound to growth inhibition and apoptosis. The present study intends to obtain additional data on the interaction of EGF with cancer cells by analyzing its effect in vitro on the growth of cultured A431 cells, and in vivo on nude mice xenotransplanted with this cell line; identifying, in addition, the gene expression patterns for a selected group of genes related to EGF and cancer pathways in the solid tumor. EGF-treated animals showed significantly smaller tumor volumes than controls, suggesting cellular growth inhibition mediated by this factor. Similar results were obtained regarding the proliferation of cultured A431 when treated with 2.2, 33 and 165 nM of EGF. These results may imply a common mechanism for the EGF-mediated growth inhibition of A431 cells in both biological conditions (in vivo and in vitro). The action of EGF at nanomolar concentrations may trigger a transient recovery of the tumor suppressor ability of tumor cells through the reduction of the biological action of mutated TP53, cell cycle arrest due to a decrease in Cdk4 expression levels and the initiation of caspase through the activation of CASP9.
Keywords: epidermal growth factor, cancer, A431 cells, nude mice.
RESUMEN
El carácter de promotor de la carcinogénesis atribuido al factor de crecimiento epidérmico (FCE) no se ha reproducido uniformemente, pues las células malignas tratadas con esta molécula en algunos experimentos han mostrado inhibición del crecimiento y apoptosis. Se ofrecen datos adicionales de la interacción del FCE con células cancerosas, mediante el análisis de su efecto en el crecimiento de la línea celular A431 in vitro y en ratones atímicos xenotrasplantados con esta línea. Se estudiaron además los patrones de expresión de un grupo de genes relacionados con el FCE y el cáncer en los tumores sólidos de A431. Este estudio reveló que los animales tratados con FCE humano recombinante (FCEhr) desarrollaron volúmenes tumorales inferiores que los animales controles. Ello sugiere una inhibición del crecimiento celular mediada por el FCE. Resultados similares se obtuvieron in vitro al tratar las células A431 con FCEhr a 2.2, 33 y 165 nM. A estas concentraciones, el FCE recobra la propiedad supresora tumoral en las células cancerosas, posiblemente por la reducción de la acción biológica de la TP53 mutada, la inhibición del ciclo celular al disminuir la expresión del gen cdk4, y la iniciación de la vía de las caspasas mediante la activación de la expresión del gen CASP9. Tales hallazgos sugieren mecanismos comunes de inhibición del crecimiento de las células A431 in vitro e in vivo, mediados por el FCE.
Palabras clave: factor de crecimiento epidérmico, cáncer, células A431, ratones atímicos.
INTRODUCTION
The epidermal growth factor (EGF) is a 53-amino acids polypeptide originally isolated from mouse salivary glands. EGF discovery was presided by its ability to stimulate epithelial growth and differentiation upon its injection to newborn mice. In extraembryonic life, the homeostatic preservation of epithelial cell populations by controlling cell growth, proliferation, and differentiation is one of its most remarkable physiological roles [1, 2].
It is likely that Stanley Cohen was the first to exogenously administer EGF in pharmacological concentrations to an animal model, thus evoking an expected clinical response while healing corneal burns in rabbits. This opened up the first pharmacological attempt to improve the healing process using the exogenous administration of a growth factor [3].
According to years of research in different biological systems, the exogenous administration of EGF at supra-physiological concentrations triggers two major pharmacological actions: cyto-protective and trophic-reparative [4]; which are critical for the survival and repair process of internal and external organs. However, the fact that some growth factors were identified in the conditioned medium of different cancer cell lines and the existing homology between some growth factors and/or their receptors and viral oncogenes engendered an early concern within the medical community. Accordingly, the events whereby EGF and other growth factors stimulate tissue repair are similar to those involved in tumorigenesis: cell proliferation, migration, survival, and de novo angiogenesis [5].
In contrast to other ligand family members, EGF itself is not an oncogene-derived product but its specific receptor is encoded by an oncogene (EGFR, ERBB1) endowed with tyrosine kinase activity. Several EGFR mutations correlate with tumor aggressiveness and patient’s poor prognosis. Nowadays, EGFR constitutes a therapeutic target of central importance for most epithelial cancers [6, 7].
In contrast to the tumorigenic role of a deregulated EGFR, the co-carcinogenic effect of EGF appears controversial so far [7-9]. The EGF mediated-carcinogenic promotion has not proved reproducibility in vivo through different experimental settings and some data reveal an EGF-induced inhibition of several cancer cell lines of epithelial origin [10-13]. Parallely to the above basic findings, EGF emerged and progressed in the clinical arena. The yeast-made recombinant human molecule stands as a major therapeutic tool within the clinical possibilities of hard-to-heal lower extremity wounds in diabetics [14-16]. Within this realm, its intralesional administration has been demonstrated to trigger and sustain the healing process of poor-prognosis diabetic ulcers, which can reduce the rate of lower extremity amputations [14-16].
The present study is, therefore, aimed at providing further data on the impact of recombinant human EGF (rhEGF) in a broadly used human carcinoma line, examining the A431 cells proliferative response in vitro as the changes in tumor size in vivo. In addition, we studied the gene expression patterns of a selected group of genes related to EGF and cancer pathways in the solid xenografted tumors.
MATERIALS AND METHODS
Cell culture
The A431 human epithelial carcinoma cell line (donated by Dr. Belinda Sánchez from the Center of Molecular Immunology, Cuba) is a model cancer cell line over-expressing EGFR, often used in biomedical research to explore cancer pathways [10]. Cells were cultured in RPMI 1640 (Gibco-BRL®, Gaithersburg, MD, USA), supplemented with 10% fetal calf serum (PAA Laboratories, Germany), 1% L-glutamine, 50 µg/mL gentamycin (Sigma-Aldrich Corp, St Louis, MO, USA) at 37 °C in a 5% CO2 atmosphere with 95% relative humidity. The cells were expanded up to 20 × 106 cells for flow cytometry experiments and for the inoculation of nude mice.
Flow cytometry in A431 cells
The expression of EGF receptor (EGFR) in cells was quantified by flow cytometry. The cells were removed from the culture flasks using a trypsin-EDTA solution and centrifuged at 1000 x g for 5 min; the pellet was washed then with phosphate-buffered saline (PBS) and suspended at a final concentration of 3 × 106 cells/mL in PBS containing 3% Bovine serum albumin and 10 µg/mL of the humanized anti-EGFR monoclonal antibody hR3 (Center of Molecular Immunology, Cuba) [17]. Samples without the antibody were included as controls. The mixture was incubated at 22 °C for 1 h with occasional gentle agitation, after which the cells were further washed three times with PBS by centrifugation. Then, they were incubated for 30 min in the dark with goat anti-human IgG antibodies labeled with fluorescein isothiocyanate (Sigma-Aldrich Corp, St Louis, MO, USA) diluted 1:60. Following three washes with PBS by centrifugation, the samples were resuspended in 1.5 mL of PBS and analyzed in a PAS III fluorescence-activated cell sorter (FACS) machine and using the program Flomax version 2.60 (Partec; Munster, Germany).
Effects of rhEGF on the growth of A431 cells in vitro
The rhEGF was obtained from a transformed Saccharomyces cerevisiae strain at the Center for Genetic Engineering and Biotechnology (CIGB), in Havana, Cuba [18]. This rhEGF preparation, composed of a mixture of the 52 and 51 aminoacid species, has been subjected to diverse preclinical studies [19] and is the active pharmaceutical ingredient of registered healing medications for topical use in acute lesions (Hebermin®) and for intralesional infiltration in chronic diabetic foot ulcers (Heberprot-P®). The A431 cells were seeded in 24-well plates at 25 000 cells/well using RPMI 1640 (Gibco-BRL®, Gaithersburg, MD, USA), supplemented with 10% Fetal Calf Serum (PAA), 1% L-glutamine and 50 µg/mL gentamycin (Sigma-Aldrich Corp, St Louis, MO, USA) and cultured at 37 °C in a 5% CO2 atmosphere for 24 hours. After this time, the cells were washed with PBS and incubated with the same culture medium supplemented with different rhEGF concentrations (2.2, 33 and 165 nM) or EGF-free preparation (control composition: sucrose, dextran 40, disodium phosphate, sodium dihydrogen phosphate dehydrated, and water for injection). The cells were then cultured at 37 °C in a 5% CO2 atmos-phere for 72 h, after which they were removed from the wells using a trypsin-EDTA solution and counted using a hematocytometer.
Xenografts of human tumor cells into nude mice and evaluation of rhEGF administration effect
Twelve NIH nude mice (CIGB) were subcutaneously inoculated on the right dorsal region with 300 µL of RPMI 1640 medium containing 3 × 106 of A431 cells per animal. Once the tumor became visible and palpable (approximately 1-cm diameter) the mice were randomized (six animals for the rhEGF treatment group, the remaining six for control treatment). Both groups were injected near the tumor twice a week (approximately dose per kg and frequency to that used in rhEGF treatment of diabetic foot ulcers), using either 1 µg rhEGF or the equivalent volume of control solution per kg of bodyweight (in this case 20 ng of rhEGF (2.2 nM) or 100 µL of EGF-free preparation per 20 g of mice bodyweight), during 17 days. The results presented reflect the tumor growth and survival events from three experiments ex-temporarily and independently made.
The nude mice experiments were conducted under the observation of ethical procedures and the experimental protocol was reviewed and approved by the animal welfare committee of the CIGB.
Tumor size (length and width) was measured every other day, and its diameter and ratio was calculated. The volume (V) of subcutaneous tumor was estimated using the following formula V = (4/3) πr3 [20]. Once the administration scheme concluded, the animals were terminated and autopsied. Tumor samples were harvested for both: histopathology studies and to compare the gene expression profiles from the rhEGF-treated and control groups.
Histopathology
The specimens from the tumors (4 samples from treated animals and 3 samples from controls) were processed for histopathological study following routine techniques. Briefly, the samples were fixed in 10% buffered formalin, paraffin embedded, 5-µm sectioned and hematoxylin and eosin stained. Tumors were evaluated based on the following parameters: total cells in mitosis (TCM), tumor total cells (TTC), mitotic index (MI), total cells in apoptosis (TCA), apoptotic index (AI), mitosis-apoptosis ratio (MAR), total vessels neoformation (TVN) and neoformation vessels index (NVI). Ten randomly-selected fields with 100-cells each were used for quantifying all these parameters using 100x magnifications [21]. For the evaluation of these parameters the software Madip, version 4.0, and the following equations, as described Rodríguez et al. [22] were employed:

Isolation of RNA
Tumor biopsies were collected into RNAlater® (Ambion, AB applied Biosystem, USA) and stored at -20 °C for up to 2 weeks. The samples were then processed on a TissueLyser (Qiagen, Hilden, Germany); extracting total RNA with the TriReagent method (Ambion, AB applied Biosystem, USA). RNA purity and yield were determined using a NanoDrop Spectrophotometer (NanoDrop Technologies, USA), and RNA integrity number (RIN) was obtained on a Bioanalyzer analysis (Agilent, 2100, USA).
cDNA synthesis
Total RNA was treated with RQ1 RNase-Free DNase (Promega corporation, Fitchburg, Wisconsin, USA) in order to remove any DNA contaminants from the preparation. First strand complementary DNA (cDNA) was synthesized from 1 µg of total RNA using SuperScript™ II reverse transcriptase (Invitrogen technologies, Carlsbad, California, USA) and oligo-dT primers following fabricant´s instructions.
Selection of the gene set for Real-time-quantitative-PCR analysis
Gene expression of 49 genes selected from the EGF signaling cascade and cancer pathway (supplementary table for gene names, symbols and NCBI reference sequence transcript accession numbers) were analyzed, following the recommended signaling pathway in SABiosciences (Qiagen Company, Hilden, Germany). In order to characterize the selected set of genes on a genomic scale, software for generating molecular interaction networks and gene enrichment tools were used. The protein interaction network, composed of the direct interactions between the genes of the selected set, was generated with Cytoscape [23] using the BisoGenet plugin [24], using the whole set of databases available at the BisoGenet as primary source of protein interactions. This network was augmented by adding edges between genes coding for transcription factors and their target genes. This was done by compiling transcription regulation data from TRANSFAC® [25] (version 7.0, 2005) using its web interface [26], employing then this information to generate a Cytoscape .sif file. This file was imported into Cytoscape and merged with the protein interaction network created by using BisoGenet. Significant biological processes of Gene Ontology (GO) [27], were investigated with Cytoscape’s plugin BiNGO [28], and significant pathways were identified with DAVID [29], KEGG [30] and BioCarta [31] databases as primary data sources on pathways. In both cases, a p-value of 0.05 was chosen as threshold for statistical significance.
Real-time-quantitative-PCR
A primer3 web application [32] was used to design Real-time-quantitative-PCR (qPCR) primers with a length of 22 bases and an average GC content of 50%. GAPDH, HMBS and YWHAZ were selected and tested as reference genes, due to the stability of their transcription levels across the sample set. We employed real-time PCR to analyze gene expression in the solid tumors of nude mice xenografted with A431 cells. A total of 4 animals from the rhEGF-treated group and three from the control were included, using two technical replicates per gene. The reactions were performed in a volume of 20 μL, including 10 µL of PCR AbsoluteTM QPCR SYBR Green Mix (Thermo Scientific), 6 μL of primers (70 nM) and 4 µL of cDNA diluted by a factor of 20. The reactions were performed in an optical detection rotor for 36 tubes at 95 °C for 15 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s. qPCR data analysis was performed with CapitalBio RT-Cycler series analysis software (version 2.001, CapitalBio Co, Ltd, Beijing, China). Relative quantification to untreated control and normalization with the reference genes were performed with REST 2009 version 2.0.13 (Qiagen GmbH) [33]. The LinRegPCR program (version 11.3, 2009; Amsterdam, the Netherlands) was used to estimate PCR efficiencies [34] and geNorm was used for the selection of the most stable reference genes from a set of tested genes in a given cDNA sample panel [35].
Statistical analysis
To study if significant differences existed in the tumor volume, A431 cell proliferation, and histophatological analysis between rhEGF-treated group and control, a normality analysis (Shapiro Wilk’s test) and homogeneity of variance test (Levene’s test) were carried out. Groups were compared using the Student’s t-test (parametrical) or Mann-Whitney’s U test (non-parametrical) applied to independent samples in the in vivo study, and Student’s t test applied to independent samples in the in vitro study. In order to corroborate the results, due to small size of samples in each group, mean comparison by estimating confidence intervals or credibility to 95% and 90% by Bayesian Inference methods applied to independent samples were also done. The significance level chosen was 0.05. Statistical analyses were done using SPSS for Windows version 15.0, and EPIDAT for Windows version 3.1, Bayesian Module [36].
RESULTS
Flow cytometry
In the FACS experiments, A431 human tumor cells were incubated with the hR3 monoclonal antibody against EGFR. Mab hR3 bound to 99.3% of the cells after subtracting the signal of the negative control, thus showing the high density of EGF receptors expressed by this cell line (Supplementary figure 1). This observation confirms previous reports that the A431 cell line expresses about 2.6 × 106 molecules of EGFR per cell [10].
Effect of different concentrations of rhEGF on the proliferation of A431 cells
In order to examine the effect of rhEGF on the inhibition of A431 proliferation, cells were exposed to different EGF concentrations, being 2.2 nM, the dose used in the treatment of diabetic foot ulcers. Cells were exposed to a range of doses of 2.2, 33 and 165 nM of rhEGF during 72 h; also including rhEGF-free preparation (control) and untreated cells controls. All the assayed rhEGF concentrations inhibited A431 cells proliferation. Graphical representation of cell proliferation and the results of statistical analysis are shown in figure 1. Growth of EGF-free preparation treated cells did not differ from untreated ones.
Tumor growth in mice
Changes in the subcutaneous solid tumor mass volumes were observed between rhEGF and control treated mice. On day 15, the control-treated group had an average tumor volume of 292 ± 27 mm3, compared to 121 ± 10 mm3 for the rhEGF-treated group. Tumor growth curves revealed the existence of large significant differences between the groups treated with control and rhEGF (Figure 2). Similar results of tumor growth between animals treated with rhEGF (1 µg/kg of animal body weight) and the control group were observed in two replication of this study using a larger number of animals and time-course treatment (Supplementary figure 2).
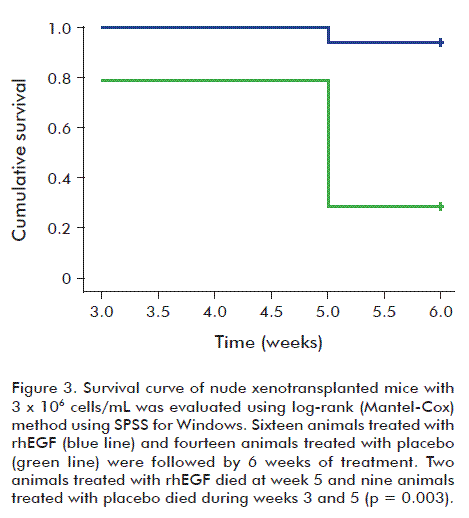
Epidermal growth factor prolongs survival time of tumor-bearing mice
It was observed that rhEGF prolonged the survival time of mice bearing A431 tumor xenografts vs. controls. Figure 3 and table 1 show the result of log-rank (Mantel-Cox) analysis in a study made using 3 × 106 cells xenotrasplanted/animal and treated with 20 ng of rhEGF or control. As shown inxenotrasplanted/animal and treated with 20 ng of rhEGF or control. As shown inxenotrasplanted/animal and treated with 20 ng of rhEGF or control. As shown in table 1, rhEGF-treated animals lived 5.87 weeks, but control animals lived 4.9 weeks, which was significantly different (p = 0.003). In this animal model, the 2.2 nM concentration of rhEGF prolonged the survival of animals one week with respect to the controls.
Histopathological study
The histopathological examination of the primary tumors, together with the calculation of the corresponding mitotic and mitotic/apoptotic indexes revealed the existence of significant differences in cell division between the groups. The mitotic index at 16 days showed that the proliferation status of cells sampled at these times had less number of cells in mitosis in relation to the total number of cells (Table 2). These indexes for the implanted tumors in the EGF-treated group were lower than in the control group.
Gene expression analysis in xenotrasplanted tumors (A431 cells) in nude mice
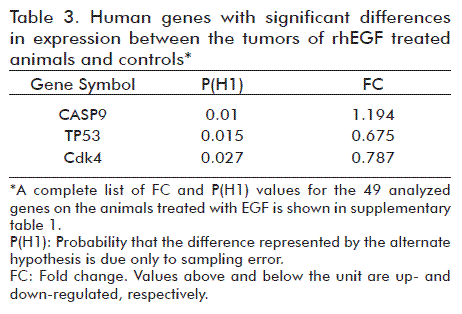
The isolated RNA appeared to meet the quality standard for further use in differential gene expression analyses by qPCR (data not shown). All the samples had RIN larger than 7. The qPCR technique was validated to demonstrate the absence of genomic DNA in the RNA preparations and the absence of inhibitors in the cDNA synthesized for the PCR reactions. A similar observation was found after the evaluation of references genes with the least variation in their expression levels between samples of rhEGF-treated group and the control. The use of qPCR addressed the identification of genes that could be involved in the observed cell proliferation inhibition in vitro and in vivo after the application of rhEGF. According to the results, 6% of the analyzed genes had significantly different expression levels between both treatment groups. Compared to the control animals, two of the three genes with significant differences had lower expression levels but the expression of the third was increased (Table 3). A molecular interaction network was designed using, as input, some of the most relevant genes that interact directly with Cdk4, TP53 and CASP9, genes that varied significantly their expression (Figure 4).
DISCUSSION
EGF is endowed with proliferative and cytoprotective abilities, promoting cell survival and tissue repair. There is a controversy about the role of EGF in cancer development because: its specific receptor is encoded by an oncogene, the EGFR is mutated in most epithelial cancers and the signals, produced by the interaction of EGF and its receptor to stimulate tissue repair such as proliferation, migration and angiogenesis, are the same signals that promote tumorigenesis [37].
Although an obvious concern still survives before the clinical use of EGF, a thorough examination of the international reported clinical interventions suggest that EGF treatments have been well tolerated (including systemic exposure) and safe within temporary biological windows enough for malignant processes’ onset (4 to 15 years of follow up) [37].
Our study has shed further light on the safety profile of EGF, which at the effective clinically concentrations does not favor the development of tumors in A431 cells. We developed an A431 tumor-bearing nude mice model and it was validated through phenotypical and histological tests. A431 is a highly tumorigenic squamous carcinoma cell line that was chosen due to its large levels of EGFR expression, which provide a large dynamic range for testing the effect of different EGF concentrations on the activation of the EGFR and cancer signaling pathways [10].
In this study, we found that concentrations of rhEGF of 2.2, 33 and 165 nM produced cell proliferative inhibition of A431 during cultivation. In other words, rhEGF concentrations up to 16-fold higher than those reported by other authors produced an inhibitory effect on growth, similar to EGF at 0.1-10 nM
[38-41]. For the in vitro study, the experimental point of 2.2 nM of rhEGF matches with the concentration used in the clinics during the therapy of chronic ulcers [14-16]. The results as a whole, suggest that rhEGF does not promote the proliferation of the A431 cell line and, therefore, may not assist in tumorigenesis in these cells.
The high levels of EGFR in A431 cells appears to facilitate tumor growth in vivo, compared other tumor lines expressing lower levels of EGFR. These observations are consistent with the results of Barnes in 1982 [40] and Santon et al. in 1986 [41], which demonstrated that the concentration of EGFRs in A431 cells is directly proportional to their ability to grow as solid tumors in host animals. In line with this, the rhEGF-treated animal group showed significantly smaller tumor volumes than the control group, suggesting an inhibition of tumor growth mediated by rhEGF. We observed a similar behavior for A431 cells growing in vitro under the influence of nanomolar concentrations of rhEGF. These results might suggest that EGF promotes similar mechanisms in both biological scenarios (in vivo and in vitro), resulting in the growth inhibition of A431 cell line.
In addition to the tumor growth data, the histological analysis also suggests that rhEGF exerts an inhibitory, rather than stimulatory mitotic action on A431 tumor cells. The mitotic index, a critical parameter for cancer prevention and control, was lower in rhEGF-treated animals compared to the control, possibly due to cell cycle arrest arising from the repair of genetic material and the promotion of tumor cell death. These could be transient events that should be promoted by the rhEGF action on A431 tumors in athymic mice [42].
The rhEGF also favored the survival of animals inoculated with 3 million A431 cells, perhaps providing protective properties against tumor aggressiveness and invasiveness. As proposed by Amagase et al., EGF may play a role in preventing the metastasis of certain malignant neoplasms, thus prolonging the survival time of athymic nude mice bearing human xenografts [43].
Tumor phenotypic descriptions are accompanied by changes produced by EGF`s molecular mechanisms of action, which generate a cascade of intracellular signals that justify its biological effect and cellular response. The gene expression profile that characterized the treated group supports the above statement through genes that varied significantly their expression under the action of rhEGF (Figure 4).
Three genes (TP53, Cdk4 and CASP9) with relevance in cell cycle regulation and apoptosis showed significant differences in expression in the tumors of EGF-treated animals vs. Control. These genes with subtle changes in their expression levels (fold-change lower than 2) could be indicating that they are more tightly regulated in its effect on the tumor biology formation [44].
The results showed that rhEGF at nanomolar concentrations inhibited the proliferation of A431 tumor cells both in vitro and in vivo. This cell line has a point mutation in the TP53273.His that contributes to malignant progression through genomic unstability due to the inhibition at the G1 arrest point [45]. TP53273.His is considered as a type I missense mutation, since it affects residues of the DNA-binding surface and disrupts the contact points between protein and DNA [46]. Some of the phenotypic effects of TP53273.His are: increase cell proliferation, increase growth density and induction of antiapoptotic activity [47]. The down-regulation of TP53 mRNA levels in tumors cells treated with rhEGF may be related with a reversible cell cycle arrest at the G1-S boundary. In this study, we observed a possible link between the effect of rhEGF and the down-regulation of two important cell cycle regulator genes, the TP53273.His and Cdk4, and their implication in the lengthening of the G1 to S phase transition, therefore inhibiting tumor growth. The results are not enough to further dissect and identify what genes or protein interactions lead to decrease gene expression levels for Cdk4, but the decrease in TP53 levels due to EGF may suggests a reduction of the oncogenic gain-of-function of the TP53 gene and a reprogramming of the activity of genes controlling the cell cycle, such as Cdk4.
Another downstream component of the EGF regulatory cascade is Caspase-9, a gene for a member of the cysteine-aspartic acid protease (caspase) family. Sequential activation of caspases plays a central role in the execution phase of cell apoptosis and therefore, the restoration of some cell pathways through the action of EGF in tumor cells may drive them directly to cell death (apoptosis) [48].
Preliminarily, in the in vivo scenario, the action of rhEGF led to a transient recovery of tumor suppressor ability in tumor cells through the reduction of the biological action of mutated TP53, the arrest of cell cycle due to decreased levels of Cdk4 and the promotion of apoptosis through the activation of CASP9.
Our findings are not surprising. They rather support previous evidences documenting an EGF-mediated anti-tumorigenic effect at certain supraphysiological concentrations in A431 cells [38-40]. Other malignant cell lines as MX-1, UM-1 of mammary origin as the ES-4 derived from esophageal cancer are also inhibited by EGF in proportion to the concentrations injected, when transplanted to athymic recipients [42]. More recent studies have shown that EGF failed to enhance in vitro proliferation of three human gastric adenocarcinoma lines, as there were no effect on tumor growth when these cells were implanted in nude mice [49]. Curiously, for both the in vitro and in vivo approaches, the experimental protocols and the EGF doses have been largely used and known to facilitate proliferation of non-transformed cells. Lately, according to recent in vitro findings EGF induced more noticeable changes in A431 cells proliferation and death than cetuximab or gefitinib. Both, cells’ arrest and death appeared far more prolonged and irreversible by EGF than that of the conventional anti-tumor drugs [50].
CONCLUSIONS
These results provide evidences in favor of the safe use of EGF in clinical practice, which at nanomolar concentrations prolongs the animal survival time and inhibited the proliferation of tumor in animal’s xenotrasplanted with A431 cells. The findings of differential gene expression in vivo analysis reinforce these phenotypic observations. Important genes for the cell cycle regulation, as TP53, Cdk4 and CASP9 varied significantly their expression levels favoring cell cycle inhibition and apoptosis.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Jorge Gavilondo for his scientific contribution to the planning and development of this research. We also want to thank Dr. José Angel Silva for his excellent dedication to the synthesis of real time PCR primers.
REFERENCES
1. Carpenter G, Cohen S. Epidermal growth factor. J Biol Chem. 1990;265(14):7709-12.
2. Venturi S, Venturi M. Iodine in evolution of salivary glands and in oral health. Nutr Health. 2009;20(2):119-34.
3. Kresge N, Simoni RD, Hill RL. Precocious Newborn Mice and Epidermal Growth Factor: the Work of Stanley Cohen. J Biol Chem. 2006;281(10):e10.
4. Berlanga J. Heberprot-P: experimental background and pharmacological bases. Biotecnol Apl. 2010;27(2):88-94.
5. Cross M, Dexter TM. Growth factors in development, transformation, and tumorigenesis. Cell. 1991;64(2):271-80.
6. Gazdar AF. Personalized medicine and inhibition of EGFR signaling in lung cancer. N Engl J Med. 2009;361(10):1018-20.
7. Berlanga J, Gavilondo J, García del Barco D, Martín J, Guillén G. Epidermal Growth Factor (EGF) and Platelet-Derived Growth Factor (PDGF) as Tissue Healing Agents: Clarifying Concerns about their Possible Role in Malignant Transformation and Tumor Progression. J Carcinogenesis Mutagenesis. 2011;2(1):115. Available from: http://www.omicsonline.org/2157-2518/2157-2518-2-115.php?aid=294.
8. Berlanga J, Álvarez S, de la Fuente J, López P. Considerations on the transforming potential of the epidermal growth factor. Biotecnol Apl. 1998;15(2):65-9.
9. Berlanga-Acosta J, Gavilondo-Cowley J, López-Saura P, González-López T, Castro-Santana MD, Lopez-Mola E, et al. Epidermal growth factor in clinical practice - a review of its biological actions, clinical indications and safety implications. Int Wound J. 2009;6(5):331-46.
10. Stoscheck CM, Carpenter G. Biology of the A-431 cell: a useful organism for hormone research. J Cell Biochem. 1983;23(1-4):191-202.
11. Masui H, Castro L, Mendelsohn J. Consumption of EGF by A431 cells: evidence for receptor recycling. J Cell Biol. 1993;120(1):85-93.
12. Jakus J, Yeudall WA. Growth inhibitory concentrations of EGF induce p21 (WAF1/Cip1) and alter cell cycle control in squamous carcinoma cells. Oncogene. 1996; 12(11):2369-76.
13. Sonoke S, Ueda T, Fujiwara K, Sato Y, Takagaki K, Hirabayashi K, et al. Tumor regression in mice by delivery of Bcl-2 small interfering RNA with pegylated cationic liposomes. Cancer Res. 2008;68(21):8843-51.
14. Acosta JB, Savigne W, Valdez C, Franco N, Alba JS, del Río A, et al. Epidermal growth factor intralesional infiltrations can prevent amputation in patients with advanced diabetic foot wounds. Int Wound J. 2006;3(3):232-9.
15. Fernández-Montequín JI, Infante-Cristia E, Valenzuela-Silva C, Franco-Pérez N, Savigne-Gutiérrez W, Artaza-Sanz H, et al. Intralesional injections of Citoprot-P (recombinant human epidermal growth factor) in advanced diabetic foot ulcers with risk of amputation. Int Wound J. 2007;4(4):333-43.
16. Fernández-Montequín JI, Valenzuela-Silva CM, Díaz OG, Savigne W, Sancho-Soutelo N, Rivero-Fernández F, et al. Intra-lesional injections of recombinant human epidermal growth factor promote granulation and healing in advanced diabetic foot ulcers: multicenter, randomised, placebo-controlled, double-blind study. Int Wound J. 2009;6(6):432-43.
17. Talavera A, Friemann R, Gómez-Puerta S, Martínez-Fleites C, Garrido G, Rabasa A, et al. Nimotuzumab, an antitumor antibody that targets the epidermal growth factor receptor, blocks ligand binding while permitting the active receptor conformation. Cancer Res. 2009;69(14):5851-9.
18. Cinza AM, Quintana M, Lombardero J, Poutou R, Pérez E, Pérez LC, et al. Establecimiento de un cultivo discontinuo para la producción del factor de crecimiento epidérmico humano en levaduras. Caracterización del producto. Biotecnol Apl. 1991;8(2):166-74.
19. Berlanga-Acosta J, Playford RJ, Mandir N, Goodlad RA. Gastrointestinal cell proliferation and crypt fission are separate but complementary means of increasing tissue mass following infusion of epidermal growth factor in rats. Gut. 2001;48(6):803-7.
20. Toso C, Trotter J, Wei A, Bigam DL, Shah S, Lancaster J, et al. Total tumor volume predicts risk of recurrence following liver transplantation in patients with hepatocellular carcinoma. Liver Transpl. 2008;14(8):1107-15.
21. Leung TW, Xue WC, Cheung AN, Khoo US, Ngan HY. Proliferation to apoptosis ratio as a prognostic marker in adenocarcinoma of uterine cervix. Gynecol Oncol. 2004;92(3):866-72.
22. Rodríguez R, Alarcón TE, Sáchez LB. MADIP: Morphometrical Analysis by Digital Processing. In: Sánchez JS, Pla F. Proceedings of the IX Spanish Symposium on Pattern Recognition and Image Analysis: Benicasim (Castellón), Spain, 16-18 May 2001. Vol. I. Castellón de la Plana: Publicaciones de la Universitat Jaume; 2001. p. 291-8.
23. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-504.
24. Martín A, Ochagavía ME, Rabasa LC, Miranda J, Fernández-de-Cossio J, Bringas R. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinformatics. 2010;11:91.
25. Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31(1):374-8.
26. Public Databases for Academic and Non-profit Organizations [Internet]. Wolfenbüttel: BIOBASE Biological Databases. c2000-2001 - [cited 2011 Oct 14]. Available from: http://www.gene-regulation.com/pub/databases.html.
27. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25-9.
28. Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005; 21(16):3448-9.
29. Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3.
30. Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30(1):42-6.
31. BioCarta [Internet]. San Diego: BioCarta LLC; c2012 - [cited 2011 Oct 14]. Available from: http://www.biocarta.com.
32. Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365-86.
33. Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36.
34. Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37(6):e45.
35. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034.
36. Bernardo JM, Smith AFM. Bayesian Theory. Chichester: Wiley; 1994.
37. Zhang H, Berezov A, Wang Q, Zhang G, Drebin J, Murali R, et al. ErbB receptors: from oncogenes to targeted cancer therapies. J Clin Invest. 2007;117(8):2051-8.
38. Gill GN, Lazar CS. Increased phosphotyrosine content and inhibition of proliferation in EGF-treated A431 cells. Nature. 1981;293(5830):305-7.
39. Gill GN, Buss JE, Lazar CS, Lifshitz A, Cooper JA. Role of epidermal growth factor-stimulated protein kinase in control of proliferation of A431 cells. J Cell Biochem. 1982;19(3):249-57.
40. Barnes DW. Epidermal growth factor inhibits growth of A431 human epidermoid carcinoma in serum-free cell culture. J Cell Biol. 1982;93(1):1-4.
41. Santon JB, Cronin MT, MacLeod CL, Mendelsohn J, Masui H, Gill GN. Effects of epidermal growth factor receptor concentration on tumorigenicity of A431 cells in nude mice. Cancer Res. 1986;46(9):4701-5.
42. Murayama Y. Growth-inhibitory effects of epidermal growth factor on human breast cancer and carcinoma of the esophagus transplanted into nude mice. Ann Surg. 1990;211(3):263-8.
43. Amagase H, Tamura K, Okuhira M, Kakimoto M, Amano H, Hashimoto K, et al. Epidermal growth factor prolongs survival time of tumor-bearing mice. Jpn J Cancer Res. 1990;81(5):495-500.
44. Jones LB, Bean R, McLachlan GJ, Zhu JX. Mixture models for detecting differentially expressed genes in microarrays. Int J Neural Syst. 2006;16(5):353-62.
45. Williams AC, Miller JC, Collard TJ, Bracey TS, Cosulich S, Paraskeva C. Mutant p53 is not fully dominant over endogenous wild type p53 in a colorectal adenoma cell line as demonstrated by induction of MDM2 protein and retention of a p53 dependent G1 arrest after gamma irradiation. Oncogene. 1995;11(1):141-9.
46. van Oijen MG, Slootweg PJ. Gain-of-function mutations in the tumor suppressor gene p53. Clin Cancer Res. 2000; 6(6):2138-45.
47. Prasad KA, Church JG. EGF effects on p53 in MDA-468 human breast cancer cells: implications for G1 arrest. Cell Prolif. 1997;30(2):81-94.
48. Shinoura N, Sakurai S, Asai A, Kirino T, Hamada H. Caspase-9 transduction overrides the resistance mechanism against p53-mediated apoptosis in U-87MG glioma cells. Neurosurgery. 2001;49(1):177-86.
49. Xia L, Yuan YZ, Xu CD, Zhang YP, Qiao MM, Xu JX. Effects of epidermal growth factor on the growth of human gastric cancer cell and the implanted tumor of nude mice. World J Gastroenterol. 2002;8(3):455-8.
50. Song JY, Lee SW, Hong JP, Chang SE, Choe H, Choi J. Epidermal growth factor competes with EGF receptor inhibitors to induce cell death in EGFR-overexpressing tumor cells. Cancer Lett. 2009;283(2):135-42.
Received in November, 2011.
Accepted in April, 2012.
Isabel A Guillén. Departamento de Farmacogenómica, Dirección de Investigaciones Biomédicas. Centro de Ingeniería Genética y Biotecnología, CIGB. Ave. 31 e/ 158 y 190, Playa, CP 10 600, La Habana, Cuba. E-mail: isabel.guillen@cigb.edu.cu.
SUPPLEMENTARY MATERIAL