Mi SciELO
Servicios Personalizados
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Biotecnología Aplicada
versión On-line ISSN 1027-2852
Biotecnol Apl vol.29 no.4 La Habana oct.-dic. 2012
REPORT
Physiological and nutritional studies of Escherichia coli and a new combination of separation methods to obtain highly pure and homogeneous plasmid DNA for gene therapy
Estudios fisiológicos y nutricionales de la Escherichia coli y la combinación novedosa de métodos de separación permiten obtener ADN plasmídico con alta pureza y homogeneidad para su uso en terapia génica
Odalys Ruiz1, Miladys Limonta1, Jorge Valdés1, Gabriel Márquez1, Michel Díaz1, Willy Frómeta1, Martha Pupo1, Dinorah Torres1, Eduardo Martínez1, Santiago Dueñas-Carrera2
1 Dirección de Desarrollo, Centro de Ingeniería Genética y Biotecnología, CIGB. Ave. 31 e/ 158 y 190, Cubanacán, Playa, CP 11600, La Habana, Cuba.
2 Sección de Hepatitis C, Departamento de Vacunas, Dirección de Investigaciones Biomédicas, CIGB.
ABSTRACT
A scalable high-cell-density Escherichia coli culture method was established to obtain pharmaceutical grade plasmid DNA (pDNA), together with an optimized purification process. The effects of several components of the medium, such as carbon and nitrogen sources that ensure bacterial nutritional needs, were studied. The operation parameters, such as temperature, shaking and aeration, were set and the optimum values of cell growth and specific pDNA productivity in culture were determined. The subsequent purification process for pharmaceutical grade pDNA was implemented, by combining RNA precipitation with ammonium sulfate and two successive chromatographic steps consisting of size exclusion chromatography and reverse phase-high performance liquid chromatography. This work comprised the first report on the use of reverse phase to purify DNA for application in humans.
Keywords: plasmid DNA, purification, Escherichia coli, cell culture, size exclusion chromatography, RP-HPLC.
RESUMEN
Se estableció un método de cultivo incrementado para obtención de ADN plasmídico en Escherichia coli a altas concentraciones celulares, y estudio el efecto de varios componentes del medio, como las fuentes de carbono y de nitrógeno para garantizar las necesidades nutricionales. Se estableció los parámetros de temperatura, agitación y aireación, y obtuvo los valores óptimos de crecimiento celular y productividad específica de ADN plasmídico en el cultivo. Se implementó un proceso productivo de ADN plasmídico de grado farmacéutico, combinando la precipitación de ARN con sulfato de amonio, y dos pasos cromatográficos sucesivos de cromatografía de exclusión molecular y de cromatografía líquida de alta presión en fase reversa. Este último proceso es la primera vez que se emplea en la literatura para la purificación de ADN para aplicación en humanos.
Palabras clave: ADN plasmídico, purificación, Escherichia coli, cultivo celular, cromatografía de exclusión molecular, HPLC-RP.
INTRODUCTION
Non-viral vectors have become an attractive genetic transfer system for possible commercial pharmaceutical products. They are also used to express specific antigens of the cell membrane and thereby stimulate and reinforce the immune system. They are a potentially favorable alternative for a new and safer generation of vaccines.
Plasmid DNA (pDNA) is an intracellular product which may be obtained with a productivity that is proportional to the final cellular density attained in culture. To obtain large amounts of biomass, the composition of the medium and the physiological conditions for microorganism growth must be considered [1].
Fermentation strategies for obtaining pDNA were recently discussed; but the effect of culture conditions on the quality of the resulting pDNA was not described. In fact, the genetic characteristics of the microorganism and the environmental variables affect the productivity of the process.
Isolation methods commonly used for pDNA in the laboratory include organic elements, mutagenic reagents, toxic components and enzymes derived from animals, which limit its industrial scale-up [2]. Because of the similarity in the chemical composition and structure of RNA and pDNA, the elimination of RNA is a challenge; mainly when considering that pDNA account for less than 1% of host cell components [3].
Therefore, this work was aimed at:
1. Studying physiological parameters of Escherichia coli, considering its elementary composition and according to the literature, to design a scalable culture medium supporting high cell concentrations.
2. Evaluating the effect of several environmental components, such as carbon and nitrogen sources to ensure the nutritional requirements of the microorganism.
3. Studying the effects of temperature, stirring and aeration on cell growth and specific productivity of the culture.
4. Identifying two orthogonal chromatographic methods to eliminate remaining contaminants in the pDNA preparation; the resulting pDNA of high purity and homogeneous, complying with requirements of regulatory agencies for injectable products administered in humans and able to be used in preclinical and clinical phase I and II studies.
This study was granted the Annual Award of the Academy of Science of Cuba in 2011.
RESULTS AND DISCUSSION
When designing a scalable culture medium it is relevant to define the amount of biomass that will be obtained, in order to fix the concentration of nutrients that must be supplied [4]. In line with this definition, a balance of culture medium was made for each nutrient, considering the concentrations described in flame emission spectroscopy studies of E. coli cells [5]. The composition of the culture medium, which is specific for the microorganism, was then established. A fed-batch 5-L fermentation culture was implemented, reaching cell densities of almost 100 g/L of wet weight and allowing DNA yields above 1.8 mg of pDNA/g of biomass [6].
Comparison of two carbon sources
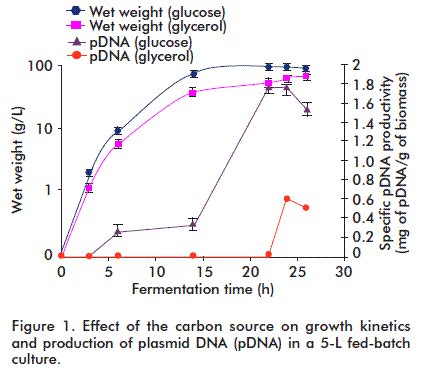
Glucose is the most widely selected carbon source because it is inexpensive and its metabolism is very efficient. However, high levels cause the undesirable production of acetate, a toxic metabolite for cells. Glycerol avoids the formation of intermediate metabolites and the accumulation of organic acids; therefore, its effect in the culture medium was considered for the production of pDNA. Nevertheless, it was observed that cell growth and specific pDNA productivity were higher when using glucose as carbon source compared with glycerol (Figure 1). Hence, the process started at low glucose concentrations in the initial medium (5 g/L), followed by slow feeding of required amounts in the scaled culture.
The appropriate composition of yeast extract maximized the production of pDNA in E. coli
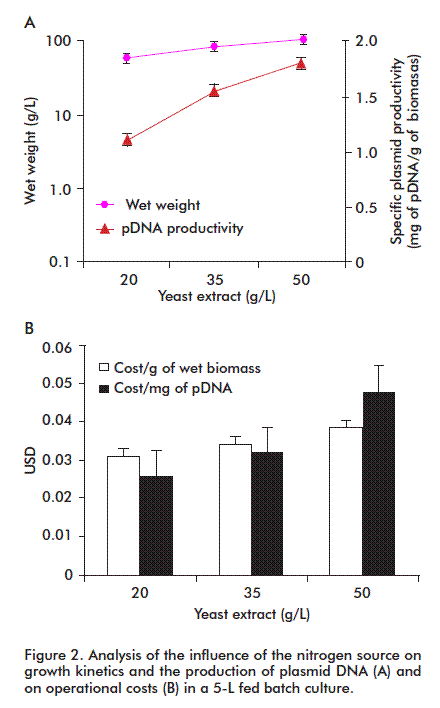
Nitrogen requirements of E. coli may be covered by adding inorganic bases, or organic as yeast extract. For this purpose, we evaluated the effect of different levels of this reagent on cell growth and specific pDNA productivity (Figure 2). Both parameters were higher at the highest yeast extract concentration (50 g/L). This concentration is affordable, with lower costs both per amount of biomass and milligram of pDNA.
An appropriate combination of temperature, shaking and aeration in the culture made it possible to recover maximum amounts of pDNA
Sometimes having the culture under sub-optimum conditions would favor a considerable increase of pDNA productivity. Therefore, the adjustment of operational parameters in a fermentation process is essential to enhance yields.
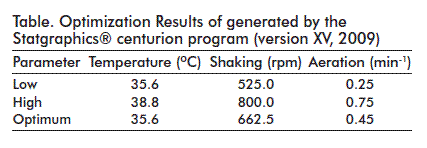
In order to determine the most favorable operational conditions, a factorial surface response Box-Behnken design was conceived with three central randomized points in which the effect of the following parameters were studied in 15 experimental runs: temperature, shaking and impellent shaking speed. Statistics were analyzed with the aid of the Statgraphics® centurion program (version XV, 2009).
A maximum local value of specific pDNA productivity (qp) was found within the interval analyzed, around the central point of the surface response generated by the model (Figure 3). The optimum value for this variable was of 264.496 mg of DNA per g of biomass, for the temperature, shaking and aeration parameters shown on the table.
Under these conditions, the large scale production of the plasmid pIDKE2 was satisfactory, on 50-L fermentations at a scaling criterion of constant weight/volume ratio during fed-batch. There were no significant differences in plasmid growth and productivity between small and large scale bioreactors [1].
Purification stage
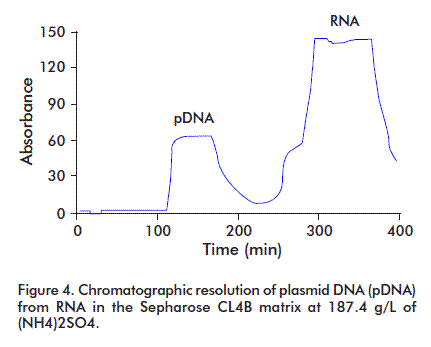
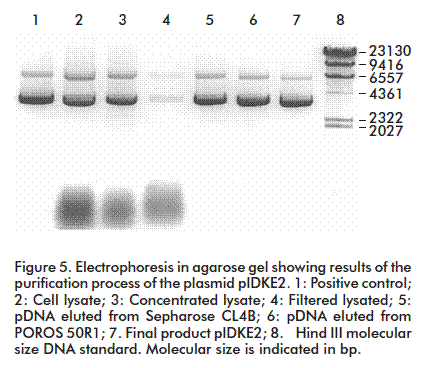
During pDNA purification, RNA may be eliminated in three ways: with RNAse treatment, by selective precipitation with Ca2+, NH4+ or Mg2+ ions [7], or by adding polyethylenglycol [8]. Here, the variation of the hydrodynamic properties of pDNA with several concentrations of (NH4)2SO4 were studied. The pDNA containing solution was applied on a Sepharose matrix CL4B column and a 91% recovery of the pDNA was observed, with a complete resolution of pDNA from RNA. The best condition was obtained at 187.4 g/L (NH4)2SO4, attaining a complete resolution of pDNA from contaminant RNA (Figure 4).
The high saline concentration affects the DNA double strand because of acting on the hydrodynamic structure of the nucleic acid molecules, either DNA or RNA. The pDNA hydrodynamic size duplicates. Therefore, at the first purification stage using size exclusion chromatography, there was a difference in the elution time between the pDNA and the RNA, and the elution volume increased (Figure 4) [3].
The second step is conceived as a concentration stage, which also removes contaminants that still remain in the pDNA containing solution. The use of the POROS 50R1 reverse phase-high pressure liquid chromatography matrix for pDNA purification is a novel strategy, based on the hydrophobic properties of the pDNA molecule, which was purified with 92% of putiry (Figure 5). The matrix showed a dynamic binding capacity of 3-5 mg/mL, which may be considered high if considering that pDNA has a large molecular mass, with no similar previous reports for conventional matrices. Another advantage of applying this matrix is the operation flow that makes it possible to work at a high speed without affecting the resolution while separating the material of interest from contaminants. This is possible because the mass transfer occurs through convective transport, a new approach to reduce limitations of mass transfer in chromatography. This leads to a drastic decrease in separation times and increases yields and productivity for pDNA recovery.
Animals immunized with the pIDKE2 plasmid produced by the methodology described in this work successfully develop antibodies against the encoded hepatitis C virus antigens.
Taking into account these results, the laboratory process was scaled up 4.5 times to obtain the pIDKE2 plasmids for preclinical and clinical studies. Lot release followed the regulations established for pharmaceutical grade DNA products [9]. Pre-clinical studies showed no systemic alterations that would jeopardize the safety of patients. Therefore, there was an adequate safety framework for the use of this product in clinical trials [10].
RELEVANCE OF THE STUDY
The methodology developed consists of the combined use, for the very first time, of the size exclusion chromatography and reverse phase-high pressure liquid chromatography for pharmaceutical grade pDNA purification. The novel use of the hydrodynamic and hydrophobic properties of the pDNA allowed purifying it with up to 92% of purity and pyrogen-free, innocuous, and sterile through a scalable, reproducible and robust process. The process does not use reagents that may be harmful for human health or the environment.
This is also the first time in Cuba that a plasmid intended for gene therapy and immunization in humans was purified with pharmaceutical grade and at preparative scale for clinical trials, in this case the pIDKE2 construct encoding hepatitis C virus antigens.
REFERENCES
1. Ruiz O, Limonta M, Valdes J, Pupo M, Martínez E. Scalable technology to produce pharmaceutical grade plasmid DNA for gene therapy. In: Kang C, editor. Gene Therapy - developments and future perspectives; Rijeka: Intech Europe; 2011. p. 53-70.
2. Diogo MM, Queiroz JA, Monteiro GA, Martins SA, Ferreira GN, Prazeres DM. Purification of a cystic fibrosis plasmid vector for gene therapy using hydrophobic interaction chromatography. Biotechnol Bioeng. 2000;68(5):576-83.
3. Limonta M, Márquez G, Rey I, Pupo M, Ruiz O, Amador-Cañizares Y, et al. Plasmid DNA recovery using size-exclusion and perfusion chromatography. Biopharm Int. 2008;21(9):38-47.
4. Yee L, Blanch HW. Recombinant trypsin production in high cell density fed-batch cultures in Escherichia coli. Biotechnol Bioeng. 1993;41(8):781-90.
5. Piatkin K, Krivoshein Y. Microbiología. Moscú: Mir; 1986.
6. Ruiz O, Pérez M, Pupo M, Limonta M, Torres D, Martínez S, et al. High-cell-density culture to produce plasmid DNA for gene therapy in E. coli. Biopharm Int. 2009;22(7):40-5.
7. Ferreira GN, Cabral JM, Prazeres DM. Development of process flow sheets for the purification of supercoiled plasmids for gene therapy applications. Biotechnol Prog. 1999;15(4):725-31.
8. Horn NA, Meek JA, Budahazi G, Marquet M. Cancer gene therapy using plasmid DNA: purification of DNA for human clinical trials. Hum Gene Ther. 1995;6(5):565-73.
9. Food and Drug Administration (FDA). Guidance for Industry: Considerations for Plasmid DNA vaccines for Infectious Disease Indications. Rockville, MD: FDA; 2005 Feb.
10. Limonta M, Márquez G, Pupo M, Ruiz O. The purification of plasmid DNA for clinical trials using membrane chromatography. Biopharm Int. 2010;23(2):46-54.
Odalys Ruiz. Dirección de Desarrollo, Centro de Ingeniería Genética y Biotecnología, CIGB. Ave. 31 e/ 158 y 190, Cubanacán, Playa, CP 11600, La Habana, Cuba. E-mail: odalys.ruiz@cigb.edu.cu.