Introduction
Coronaviruses (CoVs) are a family of enveloped RNA viruses widely distributed among mammals and birds, causing mainly respiratory or enteric diseases. However, in some cases, it is possible to observe the development of neurological diseases or hepatitis.1 CoVs have been the causative agents of two large-scale pandemics in the last two decades: 1) severe acute respiratory syndrome (SARS) in 2002 and 2003 in Guangdong Province, China; 2) Middle East respiratory syndrome (MERS) in 2012 in Middle Eastern countries.2 SARS-CoV-2, a newly identified β-coronavirus, is the causative agent of COVID-19 disease affecting the lower respiratory tract, with the potential to spread and cause damage in other organs and tissues.2,3 SARS-CoV-2 was first reported in China in December 2019, where patients affected by the virus were observed to have clinical symptoms such as dry cough, fever, dyspnea, and bilateral pulmonary infiltrates on imaging examinations.2,4
Because of the pandemic progression, people found themselves in a situation of fear and uncertainty.4 The effort to develop an effective vaccine has rendered conventional methods ineffective due to time-consuming antigen identification; lack of antigenic diversity; extensive cultivation of pathogens in laboratories; and high costs.5 On the other hand, using modern technologies in the biological sciences generates a considerable amount of data and causes a significant impact on different branches of life sciences.6
Bioinformatics is an interdisciplinary field that uses computer simulation methods to analyze biological data and make predictions about gene regulatory networks.5 This methodology has a significant advantage over conventional vaccinology techniques, including faster results and lower costs.7 Immunoinformatics is a branch of bioinformatics, a discipline whose primary goal is to convert large-scale immunological data, using computational and mathematical approaches, to understand and organize these data into immunologically meaningful interpretations.5,7 Immunoinformatics assists in the identification of possible epitopes with antigenic potential capable of generating an immune response to B and T cells.6 Elucidation of the importance of these epitopes has led to the discovery of many unknown antigens, which are a successful pathway for vaccine development.5,6 The already sequenced SARS-CoV-2 genome enables widely available computational tools and databases to aid in predicting potential B and T cell epitopes for vaccine design, immunity protein analysis, and immunization modeling.5
Bibliometrics is the study of the quantitative aspects of the production, dissemination, socialization, and disclosure of recorded information.8 It can be considered a study of research trends on a theme and thus serve as a guide for researchers to understand which subjects are mostly covered, the leading authors in the area, the main sources of information, and the most impactful countries. This tool can help qualify the research, allowing it to be more referenced and, in turn, increase the impact on the results achieved. In a proposed bibliometric analysis aimed at showing trends in vaccine research for COVID-19 using the instrument of bibliometrics, the authors concluded that the number of research studies has increased over time and that most papers have come from developed countries;4 in another analysis, with similar parameters, the authors concluded that the research foci are concentrated on vaccine side effects and public attitudes toward vaccination.9
Thus, this study aims to analyze the global trends and contributions in immunoinformatics applied to the development of SARS-CoV-2 vaccine prototypes through a comprehensive bibliometric approach.
Methods
The scientific production on the use of immunoinformatics in SARS-CoV-2 vaccine production was examine bibliometrically. For this purpose, a series of bibliometric data were collected from the Web of Science, analyzed by VOSviewer software, and thus tabulated, mapped, and graphically presented, namely: [i] the global quantity of scientific articles, identifying the most productive countries; [ii] most productive organizations/institutions; [iii] most influential authors and documents; [iv] reference co-citation network; [v] co-authorship network between countries; [vi] keyword network.
The Web of Science database was chosen as a source of retrieval material since it makes its information available in a complete way when exporting its data to tables. (10 In addition, it has a wide range of customizations in the export templates, which allows for better analysis and validation of the retrieved data.
Comprehensively, we included: articles, early access, review articles, and editorial materials that addressed the use of immunoinformatics in the production of a prototype vaccine against SARS-CoV-2. To avoid negative bias caused by the constant updating of documents in the database, all the evaluation and downloading of the material was done on a single day, November 23, 2022. Accordingly, the screening was done in the "advanced search" field, searching for the following terms in the topic field (TS): "COVID-19" or "SARS-COV-2" or "Coronavirus" and "Immunoinformatics" and "Vaccine". There were no restrictions on languages. However, the selection was made regarding the year of publication, which was restricted to the period between 2020 and 2022.
The material was retrieved in the Web of Science export option in tab-delimited format. In the "personalized" category were selected all 29 fields belonging to the groups author, title, source, abstract, keyword, addresses, references cited and use, funding and other.
In processing the extracted data, a rigorous approach was adopted to handle duplicate records. This involved a systematic screening process where titles, abstracts, and author details were cross-examined. Any duplications encountered were carefully evaluated and removed. This elimination of redundant records was crucial in maintaining the integrity of the data and ensuring the accuracy of the bibliometric analysis.
For the analysis and creation of the network maps, we used the software VOSviewer 1.6.18. The tables and graphs were made in Google Spreadsheets. The flowchart was assembled using free templates available on the Canva portal.
To avoid the occurrence of synonyms in the keyword network, a "thesaurus" file was created providing the commands for substituting words of similar meaning as follows: the terms “antibody” and “antibodies” were unified to "antibody"; “epitope” and “epitopes” were grouped together to "epitope"; “multi-epitope” and “multi-epitope vaccine” to "multi-epitope"; “coronavírus", “coronaviruses”, “sars coronavírus", “sars-coronavirus”, “human coronavirus”, “COVID-19”, “sars”, “sars-cov”, “sars-cov-2” and “respiratory syndrome coronavirus” were replaced by "sars cov 2"; similarly the terms “peptide” and “peptides” were changed to "peptide"; as were the terms “spike protein”, “spike glycoprotein” and “spike” were standardized to "spike protein".
Documents that did not have the information of the year of publication, country of origin, affiliation, and author's keywords were edited to acquire it, replacing the gaps manually in the file delimited by tabulation based on the original work of the authors; however, some of them did not have keywords but were considered for the study. Cross-referencing the information generated by the software, together with the information retrieved in tables, revealed the primary reference authors - those with the highest number of co-cited documents - from each cluster, and the DOI was used to locate their corresponding titles. Journal Citation Reports was used to calculate the impact value of the journals based on the Journal Impact Factor (JIF) for the year 2021, available on the platform.
Results and discussion
Number of publications
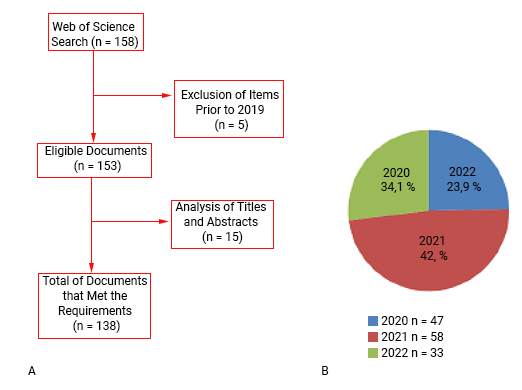
The initial search was configured to include documents published within the timeframe of 2020 to 2022. This period was chosen strategically, as it corresponds to the post-emergence phase of SARS-CoV-2, ensuring the relevancy of the data to the current research landscape. A total of 158 documents were retrieved. After analysis of titles, abstracts, and year of the study, 20 items were rejected for not discussing the topic, leaving 138. A flowchart on the study selection process is provided in figure 1a.
Publication year
An indispensable analysis in bibliometric studies, the data on publication years may be related to several other pieces of information, such as the average number of publications, most productive countries, most cited authors, and others. However, we will stick only to the total number of documents published each year. Of the 138 documents, it was found that the year 2021 was the most productive, with 58 (42.02%) records, followed by the year 2020 with 47 studies (34.06%) and 2022 with 33 (23.91%). Similarly, Wei and others used bibliometric analyzes and visualizations to document trends in COVID-19 vaccine research and found an upward trend in research publications, with a peak in October 2021. (11
Despite the increase in the number of studies in 2021, it is not possible to affirm that there is a trend of growth over time because in the following year, there was a drop in the number of publications (fig. 1b).
Publication Countries/Regions
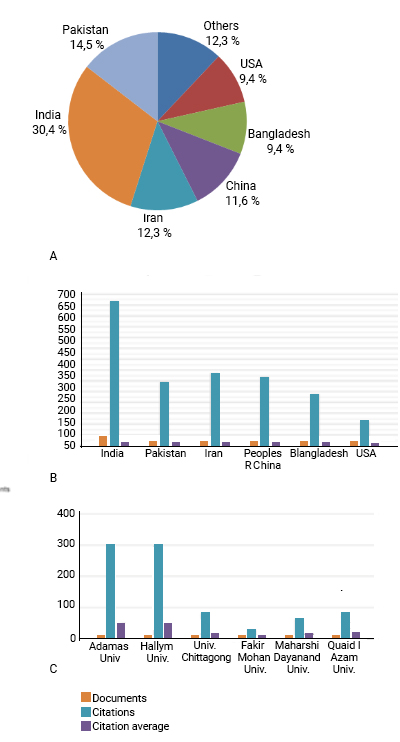
From a total of 45 countries, India was the most productive with 42 (30.4%) published papers, followed by Pakistan with 20 (14.5%), Iran with 17 (12.3%), China with 16 (11.6%), Bangladesh (9.4%) and USA with 13 (9.4%) (fig. 2a). These six nations account for more than 85% of all publications related to the use of immunoinformatics in developing a prototype SARS-CoV-2 vaccine.
Of these, India has the highest number of citations in its papers, 668, and an average of 15.90 per paper. Iran was referenced 338 times and had an average of 19.88 citations, followed by China with 319 citations and an average of 19.94, and then Pakistan was mentioned 295 times and had 14.75 citations per paper. Bangladesh was referenced 239 times, and the USA was indicated 114 times, possessing the lowest average of citations, 8.77, among these countries (fig. 2b). The country with the highest number of citations per document was Denmark, with 88.00, however, having only one document.
 Source: Own elaboration.
Source: Own elaboration.Fig. 2 - (A) Chart Countries/regions of publication; (B) Chart of the 6 most productive countries; (C) Chart 6 most productive institutions.
The study of the most productive countries is highly significant since it is representative of the funding capacity of a nation's institutions. An impact analysis shows that developed countries have more volume and relevance in published documents than developing countries but that these countries have increased the number of papers posted and have improved the impact of their articles, however, the quantitative growth of publications is not linked to the qualitative growth.12 This fact becomes more evident when we analyze the production of India concerning China, where, although India is the most productive country than China, it is China that has the highest average number of citations.
A thorough bibliometric investigation into the advancements of COVID-19 research, with a focus on vaccine safety and not exclusively relying on immunoinformatic tools, revealed a significant citation impact. (13 The publications within this domain averaged 12.14 citations each, underscoring their relevance and influence in the scientific community. The analysis further highlighted that developed countries were major contributors, accounting for 33.75% of the total research output in this field. Among these, the United States, China, and India emerged as the predominant contributors, illustrating their pivotal roles in advancing research on COVID-19 and vaccine safety. This data reflects the global effort in tackling the pandemic, with a notable emphasis on the contributions from leading nations in scientific research.
Institutions involved in publication
Complementing the country analysis, the study of institutions allows the visualization of the leading research entities of a country and their influence in the global scenario.
As highlighted earlier, India's prominent position as the most productive country, especially through its institutions such as Adamas and Fakir Mohan universities, is in line with the growing trend of emerging economies making a significant contribution to global scientific research. This finding resonates with the observations made by Chen and others in their bibliometric study, which also identified India as a key player in COVID-19 vaccine research. (14 However, our study extends this insight by highlighting the specific institutional contributions within these countries, providing a more granular view of the research landscape.
The most active institutions were Adamas University and Hallym University, each with 6 papers, obtaining 301 citations and average citations of 50.17, followed by Chittagong University with 5 publications, 82 allusions, and an average of 16.40. Fakir Mohan University, Maharshi Dayanand University, and Quaid-I-Azam University have 4 records each, which were referred 29, 62, and 81 times with average citations of 7.25, 15.50, and 20.25, respectively (fig. 2c). Guilan University of Medical Sciences was the institution with the highest average number of citations, 140.00, however, it has only one published paper. Among the 20 most productive institutions, Kangwon National University has the highest average number of citations, 95.00, having only 3 published papers (table 1).
Table 1 - The 20 most productive institutions
| Adamas Univ | 6 | 301 |
| Hallym Univ | 6 | 301 |
| Univ Chittagong | 5 | 82 |
| Fakir Mohan Univ | 4 | 29 |
| Maharshi Dayanand Univ | 4 | 62 |
| Quaid I Azam Univ | 4 | 81 |
| Shahjalal Univ Sci and Technol | 4 | 50 |
| Univ Hyderabad | 4 | 37 |
| Vidyasagar Univ | 4 | 258 |
| Bgc Trust Univ Bangladesh | 3 | 77 |
| Cent Univ Rajasthan | 3 | 24 |
| Hasanuddin Univ | 3 | 62 |
| Jahangirnagar Univ | 3 | 44 |
| Kangwon Natl Univ | 3 | 285 |
| King Khalid Univ | 3 | 30 |
| Pasteur Inst Iran | 3 | 22 |
| Shiraz Univ Med Sci | 3 | 33 |
| Univ Delhi | 3 | 29 |
| Univ Karachi | 3 | 44 |
| Univ Okara | 3 | 22 |
Source: Own elaboration.
Publishing titles
Category selection by the Web of Science allows articles with more than one research area to be included. Molecular biochemistry, immunology, and research in experimental medicine were the main research fields. Bhattacharya has the most cited paper with 226 mentions and the tenth most referenced title. Tahir Ul Qamar, too, was another author with two papers among the most cited; their publications have 105 citations. Enayatkhani has the second most cited paper, referenced 140 times (table 2). (2
Table 2 - The top 10 authors, their papers, and journals
| Bhattacharya15 | Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach | Journal of Medical Virology | 226 | 20.693 |
| Enayatkhani16 | Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in-silico study | Journal of Biomolecular Structure and Dynamics | 140 | 5.235 |
| Panda17 | Structure-based drug designing and immunoinformatics approach for SARS-CoV-2 | Science Advances | 88 | 14.980 |
| Abdelmageed18 | Design of a Multiepitope-Based Peptide Vaccine against the E Protein of Human COVID-19: An Immunoinformatics Approach | BioMed Research International | 76 | 3.246 |
| Dong19 | Contriving Multi-Epitope Subunit of Vaccine for COVID-19: Immunoinformatics Approaches | Frontiers in Immunology | 72 | 8.787 |
| Samad20 | Designing a multi-epitope vaccine against SARS-CoV-2: an immunoinformatics approach | Journal of Biomolecular Structure and Dynamics | 67 | 5.235 |
| Yang21 | An in silico deep learning approach to multi-epitope vaccine design: a SARS-CoV-2 case study | Science Reports | 59 | 4.997 |
| Tahir Ul Qamar22 | Designing of a next generation multiepitope based vaccine (MEV) against SARS-COV-2: Immunoinformatics and in silico approaches | PLOS ONE | 54 | 3.752 |
| Tahir Ul Qamar23 | Reverse vaccinology assisted designing of multiepitope-based subunit vaccine against SARS-CoV-2 | Infectious Diseases of Poverty | 51 | 10.485 |
| Bhattacharya24 | Immunoinformatics approach to understand molecular interaction between multi-epitopic regions of SARS-CoV-2 spike-protein with TLR4/MD-2 complex | Infection Genetics and Evolution | 46 | 4.393 |
Source: Own elaboration.
Reference Co-Citation Network
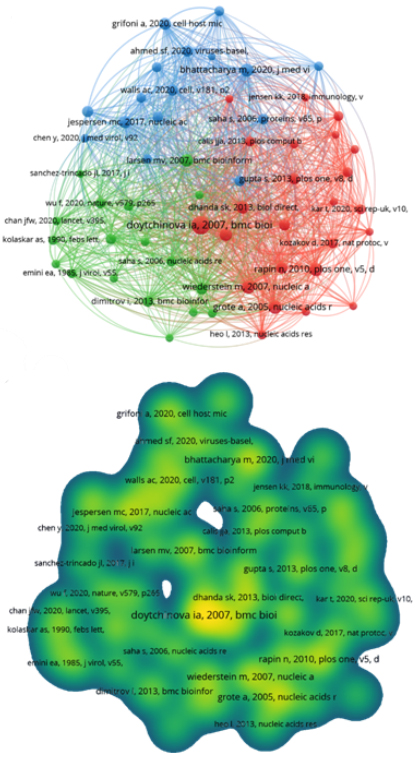
Reference co-citation analysis makes it possible to visualize the primary sources in common among authors. This analysis helps an author to publish his work in a reputable journal, and the reader can easily find the best source of information for his work. (25 In order to improve the quality of the visualization of the network maps, the minimum number of citations of a reference was set at 15. Therefore, of the 5118 publications mentioned, 58 fell within the parameters. Three clusters were generated; the first included 24 items emphasizing Doytchinova with the article "VaxiJen: a server for prediction of protective antigens, tumor antigens and subunit vaccines". The second cluster comprised 18 publications and had Larsen as the author in evidence with the title "Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction". With 42 citations, second only to Doytchinova, Bhattacharya is the author representing the highest point in the third cluster that has 16 items, with the article "Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach" (fig. 3a,b).
Inter-country co-authoring network
The analysis of co-authoring countries reflects the degree of collaboration between researchers from different countries.25,26 The minimum number of papers was set at 1, and the lowest volume of citations was recorded at 5, so only the most influential nations were selected; only 39 met the requirements.
The countries were divided into 12 clusters, of which those between the eighth and twelfth have no co-authored publications; they are Finland, Israel, Sudan, Thailand, and Turkey. The country with the highest co-publication number was Pakistan, with 15 papers. In the points in red, Saudi Arabia was the most participative of the first cluster, having 12 items in co-authorship. In the second cluster, England overlaps, with 11 records in collaboration. India has 9 links with other countries, being the most influential in the fourth cluster, followed by Malaysia with 8 co-participations, being the most prominent in the sixth cluster. The USA was the most collaborative of the light blue cluster, with 6 co-participations. The Czech Republic was the most participatory in the seventh cluster, having 4 items co-authored with other territories (fig. 4).
Considering the demonstrated collaborative efforts among countries in COVID-19 vaccine research, it's imperative to strategize for enhancing international collaboration further. 14 Key strategies include establishing global consortia for sharing data and resources, essential in fostering a unified approach to vaccine development. Standardizing research protocols through international agreements can ensure consistency and comparability in findings across borders. Joint funding initiatives are pivotal for supporting large-scale vaccine research projects, especially in resource-limited settings. Promoting partnerships between academic institutions across different countries can catalyze innovative approaches and diverse perspectives in research. Regular scientific exchanges and conferences can serve as platforms for knowledge sharing and networking among researchers worldwide. Finally, implementing policies that reduce barriers to international cooperation, such as easing restrictions on data sharing and cross-border research activities, will be fundamental in creating a more collaborative and effective global response to health crises like COVID-19. These strategies will not only aid in the current pandemic response but also lay a foundation for dealing with future global health challenges.
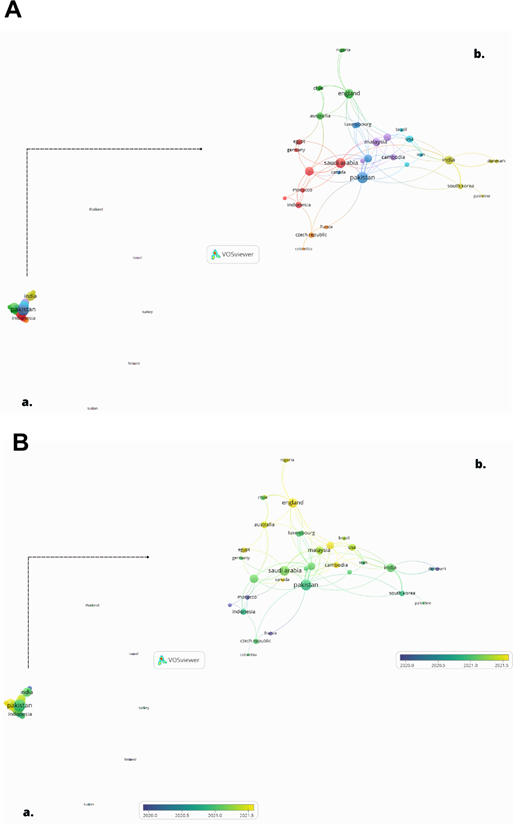 Source: Own elaboration.
Source: Own elaboration.Fig. 4 Aa - Co-authorship network between countries with studies on the use of immunoinformatics in SARS-CoV-2 vaccine development; Ab: Network of the largest node of co-authorship clusters between countries; Ba: Co-authorship time network of countries with studies related to the use of immunoinformatics in SARS-CoV-2 vaccine development; Bb: Time network of the largest node of co-authorship clusters between countries.
Keyword Network
Keywords summarize the primary information covered in the text. They contribute to greater visibility, more probabilities of citation, and a consequent increase in the impact of the scientific production of the depositing authors and the institution itself.27 In order to improve the visualization of the network maps and select only the main keywords, the minimum number of occurrences was set at 5; of the 515, only 65 met the requirements, however, due to the need for correction using the "thesaurus" file to eliminate synonyms, only 50 were analyzed.
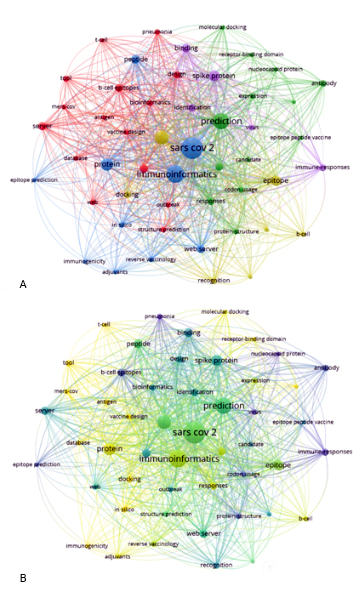
The keywords were divided into five clusters. The red dots form the first cluster and consist of 15 items with an emphasis on the terms "Server and Peptide Vaccine", occurring 18 times each. The group in green color forms the second cluster, consisting of 13 items, where the main one is the term "prediction" appearing in 71 documents. The third cluster, in blue, consists of 10 items and has "SARS CoV 2" being used 123 times. The fourth cluster is in yellow, holds 7 points, and the term that stood out was "vaccine" with frequency in 49 publications. The fifth cluster is formed by the points in lilac color, with only 5 items and having in prominence the keyword "spike protein" in 31 appearances (figure 5a,b).
Limitations
While this bibliometric analysis provides valuable insights into the field of immunoinformatics applied to the development of the SARS-CoV-2 vaccine, it encounters several limitations that must be acknowledged. First, using only the Web of Science database may overlook relevant studies published in sources that are not indexed on this platform, potentially biasing the results towards the better-known journals and excluding important papers from less prominent publications.
In addition, the methodology of collecting data on a single day (November 23, 2022) cannot fully capture the dynamics of ongoing research, as new studies are constantly appearing and existing studies receive additional citations. While this snapshot is necessary for data consistency, it may not reflect the most current state of the field.
Furthermore, it is important to emphasize that the focus on bibliometric measures such as number of publications, citations and co-authorships, while informative, does not take into account the qualitative aspects of research, such as the impact on health policy, clinical outcomes or practical applications of the findings.
Conclusion
This comprehensive bibliometric study on the utilization of immunoinformatics in developing SARS-CoV-2 vaccine prototypes has revealed several key insights, illustrating a global collaborative effort in response to the COVID-19 pandemic. The findings demonstrate a significant shift towards innovative approaches in vaccine research, underscoring the critical role of computational methods in accelerating the development process.
The analysis highlights the preeminence of countries like India, China, and the United States in spearheading research efforts, with India emerging as a significant contributor, particularly through its academic institutions. This reflects a broader trend where emerging economies are increasingly influential in the global scientific landscape. However, it is noteworthy that while India leads in terms of publication volume, China's research exhibits a higher impact factor, indicating a nuanced view of the contributions from these regions.
A striking feature of the research landscape is the extensive international collaboration, evidenced by co-authorship networks that span various countries. This trend not only facilitates the pooling of diverse expertise and resources but also enhances the potential for breakthroughs in vaccine development. The collaborative nature of the work reflects a shared global commitment to addressing the public health challenge posed by COVID-19.
The focus on the spike protein of SARS-CoV-2 as a primary antigenic target in vaccine development is a critical finding. This emphasis underscores the significance of immunoinformatics in identifying potential epitopes that can elicit a robust immune response, thus guiding the design of effective vaccine candidates.
However, the study is not without its limitations. The reliance on a single database for data retrieval, the potential for biases in citation practices, and the variability in resource distribution across different regions are factors that could influence the findings. Moreover, the rapid evolution of the COVID-19 pandemic and the emergence of new variants necessitate continuous monitoring and updating of the research landscape.
In conclusion, this bibliometric analysis serves as a testament to the power of international collaboration and innovative computational approaches in vaccine research. It underscores the need for ongoing, diverse research endeavors and the importance of adapting strategies in response to emerging challenges in the field of immunoinformatics and vaccine development.