Mi SciELO
Servicios Personalizados
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Biotecnología Aplicada
versión On-line ISSN 1027-2852
Biotecnol Apl vol.29 no.2 La Habana abr.-jun. 2012
TECHNIQUE
A microanalytical variant of the SOS Chromotest for genotoxicological evaluation of natural and synthetic products
Una variante microanalítica del SOS Chromotest para la evaluación genotóxica de productos naturales y sintéticos
Elizabeth B Cuétara1, Alba Álvarez2, Alena Alonso2, Mariolys Vernhe2, Angel Sánchez-Lamar3, Tatiana Festary1, Jeannete Rico1
1 Instituto Nacional de Oncología y Radiobiología. Esquina 29 y F, Vedado, Plaza, La Habana, Cuba.
2 Centro de Aplicaciones Tecnológicas y Desarrollo Nuclear, CEADEN. Calle 30 #502 e/ 5ta y 7ma, Miramar, Playa, AP 6122, La Habana, Cuba.
3 Facultad de Biología, Universidad de La Habana, UH. Calle 25 e/ I y J, Vedado, La Habana, Cuba.
ABSTRACT
Agents that can damage the DNA in vivo have potential adverse effects on human health. They may induce transmissible mutations and cancer. SOS Chromotest is a SOS transcriptional-fusion based assay, β-galactosidase gene was located after a SOS promoter, thus its enzymatic activity indicates the level of induction of SOS response and the DNA damage produced by chemical and physical mutagens, can be estimated. We presented and evaluated a microanalytical variant of the original SOS Chromotest for detecting genotoxicity of pigmented samples. We introduced two main modifications: we changed the colorimetric substrates for fluorescent ones and we worked at micro-analytical scale. The optimal β-galactosidase substrate concentration used was 1.8 mM and 40 minutes as time reaction. This variant detected efficiently the genotoxicity of known mutagen and the natural pigmented extracts. The results are discussed in relation to the advantages to work at micronalytical scale, costs reduction, automatization of reading and its usefulness for the screening of a large variety of samples.
Keywords: SOS Chromotest, pigmented samples, fluorescent substrate.
RESUMEN
Una variante microanalítica del SOS Chromotest para la evaluación genotóxica de productos naturales y sintéticos. Los agentes que pueden dañar al ADN in vivo tienen la potencialidad de tener efectos adversos sobre la salud humana. Los mismos pueden inducir mutaciones transmisibles y cáncer. El ensayo SOS Chromotest es un ensayo basado en una fusión transcripcional de un gen SOS, sfi A y el gen de la β-galactosidasa, así la actividad enzimática indica el nivel de inducción de la respuesta SOS response y puede estimarse el daño al ADN producido por mutágenos físicos y químicos. Nosotros presentamos y evaluamos en este trabajo una variante microanalítica del ensayo SOS Chromotest original para la detección de la genotoxicidad de muestras pigmentadas. Introdujimos dos modificaciones principales: cambiamos sustratos que desarrollan color por otros que desarrollan fluorescencia y trabajamos a escala micro-analítica. La concentración óptima de sustrato para la β-galactosidasa utilizada fue de 1.8 mM y el tiempo de reacción fue de 40 minutos. Esta variante detectó eficientemente la genotoxicidad de mutágenos conocidos y de extractos naturales pigmentados. Los resultados son discutidos en relación a las ventajas que ofrece trabajar a escala microanalítica, reducción de costos, automatización de las lecturas y su utilidad para tamizar una larga variedad de muestras.
Palabras clave: SOS Chromotest, muestras pigmentadas, sustratos fluorescentes.
INTRODUCTION
Environmental pollutants, drugs, pesticides, and natural products can damage DNA, resulting in cancers due to accumulated genetic mutations. Sensitive and simple assays have been developed for the screening and monitoring of the potential genotoxic activity of a wide range of environmental pollutants, natural and synthetic compounds. To date, the Ames test, based on the sensitivity of Salmonella strains to carcinogenic chemicals has been extensively used although certain compounds producing Ames-negative responses can in fact be carcinogenic to animals [1].
Likewise, the SOS Chromotest is a SOS transcriptional-fusion based assay capable to estimate the DNA damage produced by chemical and physical mutagens [2]. It measures the expression of a reporter gene (β-galactosidase). The β-galactosidase enzyme processes ortho-nitro-phenil galactopyranoside (ONPG) to develop a yellow compound detected at 420 nm [3]. Then, β-galactosidase induction is normalized by alkaline phosphatase activity, an enzyme expressed constitutively by Escherichia coli. The SOS Chromotest has been also widely used for genotoxicological studies [4-8]. However, this colorimetric assay has limitations when pigmented samples (e.g., phytochemical extracts) are tested, as these compounds can produce biases during the absorbance reading. Although this problem can be solved using multiple controls, it is a very laborious and time consuming procedure that introduces variability in the results.
Similar systems that solve the inconvenience of using pigmented samples have been developed for eukaryotes: a test based on yeast containing an E. coli lacZ or green fluorescent protein (GFP) reporter gene linked to the DNA damage-inducible promoter of the RAD54, RNR2 or RNR3 gene was developed [9]. In 2006, a 96-well assay system was also described based on a recombinant yeast strain containing both: a sensor and a reporter plasmid, which detects the induction of β-galactosidase activity by genotoxic compounds [10]. They also introduced three mutants defective in DNA repair and cell wall integrity in order to increase the sensitivity of the system. Podgórska et al., [11] developed a version of the Vibrio harveyi mutagenicity assay employing dark and dim mutants. V. harveyi is a naturally luminescent organism, and the test establishes that mutagen-induced reversion or pseudoreversion should cause a rescue of the ability of bacteria to emit light, a measurable characteristic. This assay was based on detection of colonies of neomycin-resistant mutants appearing frequently after a contact with mutagens being equal or higher than Ames test, depending on the nature of the tested mutagen.
In this work, we have developed a modification of the SOS Chromotest original protocol that uses fluorescent substrates, works at a microanalytical scale, and automates readings. This work was conducted: i) to define the experimental conditions of the modified assay, ii) to evaluate the sensibility of the suggested procedure and iii) to demonstrate its usefulness in the genotoxicological analysis of phytochemical extracts.
MATERIALS AND METHODS
Chemicals
The reference carcinogens (positive controls) bleomycin, cysplatin, 2-acethyl-aminofluorene (2-AAF), mitomycin C, carboplatin, doxorubycin were acquired from Nipon Kayaki, Lemery, (México), Fluka (Germany), Bioprofarma (Argentina), Korea D´New Pharmacy, and Center for Drug Research and Development (CIDEM, Cuba), respectively. Culture media were used as negative controls.
The fluorescent substrates for the β-galactosidase (4-methylumbelliferyl-β-D-galactopyranoside) and alkaline phosphatase (4-methylumbelliferyl-phosphate) assays were purchased from Sigma-Aldrich (Germany).
Bacterial strains and culture
E. coli PQ-37 strain (F- thr leu his-4 pyrD thi galE galK or galT lac ∆U169 srl300::Tn10 rpoB rpsL uvrA rfa trp::Muc+ sfiA::Mud(Ap, lac)cts) was used, as recommended for the SOS Chromotest assay [2]. Cells were grown at 37 ºC and shaken at 100 rpm in Luria-Bertani (LB) medium supplied with 50 µg/mL ampicillin, until reaching an OD600 nm= 0.4 measured in a spectrophotometer ULTROSPEC II (LKB).
Cells irradiation
In all the experiments, γ-rays (150 Gy) were used as a SOS response inducer, i.e. positive control [12-14]. Irradiation was carried out using a 60Co PX-γ-30M Russian irradiator with a temperature at 2 ± 0.5 ºC. A dose rate value of approximately 37 Gy/min was calculated using the Fricke's dosimeter [15].
Plant extracts
Aqueous extracts from the medicinal plants Phyllanthus orbicularis HBK, Pinus caribaea Morelet and Cymbopogon citratus Staff were prepared, lyophilized and conserved [15].
Modification of the original SOS Chromotest procedure
To modify the original procedure [2] the stability of the fluorescent substrate for β-galactosidase, its optimal concentration and the optimal reaction time were hereby determined. Otherwise, the alkaline phosphatase substrate (4-methylumbellipheryl-phosphate) was used at 0.43 mM dissolved in diethanolamine, as previously reported for the determination of iorT3 monoclonal antibody in UltramicroELISA [16].
First, the stability of the β-galactosidase substrate at 25 oC was tested during an hour, every 10 minutes, by measurement of the fluorescent units. Then, five β-galactosidase substrate concentrations (0.4, 0.8, 1.2, 1.8 and 2.4 mM) were tested taking as reference the original colorimetric assay [2]. Finally, the temporal course of the enzymatic activities for β-galactosidase and phosphatase alkaline were monitored every 10 minutes until one hour. All the enzymatics reactions were performed as described below.
Enzimatic reactions at microanalytical scale
Ninety-six-well microplates were used to perform the assay at microanalytical scale, so the final volume was set at 150 µL. For β-galactosidase activity detection, 13 µL of PQ-37 control and treated cells, incubated during 2 h at 37 oC, were added to 110 µL of Buffer Z (60 mM Na2HPO4, 40mM, Na2PO4, 10 mM KCl, 1 mMMgSO4, 0.1% SDS, 40 mM β−mercapto-ethanol; pH 7.0) and incubated for 20 min at 25 oC for lysis. Later, the five substrate concentrations to be tested were prepared in Buffer T (1 M Tris adjusted to pH 8.8 with HCl) and 26 µL from each one were added per well. The mixture was homogenized and allowed to react at 25 oC in darkness [2]. For alkaline phosphatase activity detection, cell lysis occurred in Buffer T and substrate was resuspended in Diethanolamine Buffer (diethanolamine 89 mM, magnesium chloride 0.13 mM, pH 9.8). In both assays, fluorescence was measured in arbitrary units (AU) using a SUMA PR-531 fluorometer (TECNOSUMA International, SA). The wavelength for substrate excitation was 365 nm and fluorescence was detected between 420 and 500 nm. Such fluorescent units were used to calculate the SOS induction factor (SOSIF) as was indicated in the original procedure [2].
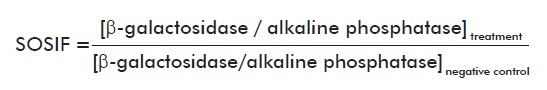
For kinetical characterization of β-galactosidase reaction, the apparent parameters maximal velocity (Vmax) and affinity constant (Km) were obtained from Lineweaver-Burk plots by using the Origin-PC package (Microcal Software. Inc).
Genotoxicity assays
Briefly, exponential phase cultures (OD600 nm = 0.4) were ten-fold diluted in fresh LB medium (2X) supplemented with ampicillin 25 µg/mL and dispensed in 1.5 mL tubes containing the compound to be tested. For each compound, the concentration range to be evaluated was set according to previous citotoxicity assays. Metabolic activation mixtures (0.4 % v/v) were used when required, to simulate the effect of hepatic enzymes in mammalians organisms. In experiments without metabolic activation the rat-liver S9 fraction was substituted by sterile distilled water. The cells were exposed to mutagens, plants extracts for 30 min at 4 oC. Later, cells were incubated for 2 hours at 37 oC and the enzymatic reactions were conducted as described above using the concentration and reaction time determined in 2.5.1.
A compound was classified as ‘not genotoxic’ if the SOSIF value remains under 1.5; as ‘not conclusive’ between 1.5 and 2.0, and as ‘genotoxic’ if it exceeds 2.0 and shows a dose-dependent correlation [17].
Statistical analysis
Data was obtained from three independent experiments, four replicas of each enzymatic activity. Mean SOSIF values, the corresponding standard deviations and normality fitness were determined for each treatment using the Kolmogorov-Smirnov test. Variance homogeneity and ANOVA tests were also conducted. SOSIF values for different treatments were compared with the control using a Dunnett test (p < 0.05) and among them using a Duncan test. For those analyses the Statistic 6.0 Software were used. For kinetical parameters determining, data fit analyses were computed in the Origin-PC package (Microcal Software. Inc).
RESULTS
Modified procedure
For the determination of the optimal β-galactosidase substrate concentration and reaction time, a physical mutagen (150 Gy of γ-rays) was used as inducer of the SOS response [14]. In the alkaline phosphatase assay, we used the buffer conditions and substrate concentration described by Salvo et al. [16]. This substrate concentration for phosphatase alkaline was tested in our conditions (data not shown).
The β-galactosidase substrate was stable during one hour at 25 °C (Figure 1). This stability corroborated the fact that the observed fluorescence increment during the assay, is due to β-galactosidase activity but not to substrate degradation. Also, 4-methylumbelliferone production depended on the reaction time (Figure 2A). Although maximum fluorescence values were reached using the substrate at a concentration of 2.4 mM, we selected 1.8 mM as the optimal concentration, since no significant increase in SOSIF values was observed when the higher substrate concentration was used (p <0.05; Figure 2B).
In order to determine an adequate reaction time for our assay, we performed readings every 10 minutes for an hour. Our intention was to reduce the original time proposed by Quillardet, but umbelliferone production was very poor in the first 30 minutes of the enzymatic reaction and started to be relevant from 40 minutes. However, by increasing reaction time in a 33% (40 to 60 min) only slightly increasing of SOSIF values (11%) was achieved. Besides, at 40 min the fluorescent signal was 35-fold and SOSIF was eight-fold the value of negative control, which is good enough for genotoxicological and antigenotoxicological studies (Figure 2B). Thus, we selected 40 min as the enzymatic reaction time, which is in accordance with that set by Quillardet in the original procedure.
The apparent Vmax and Km values were then obtained from Lineweaver-Burk plots in order to compare the affinity of the enzyme for the fluorescent substrate in the present assay with other enzymatic assays reported for β-galactosidase. The apparent Km value found was 1.9 mM for β-galactosidase in the presence of the fluorescent substrate 4-methylumbelliferyl galactopyranoside (Figure 2C).
Genotoxicity assays
To evaluate the sensibility of the modified protocol in the detection of genotoxic activity different mutagens and plant extracts were tested.
As expected, all the reference mutagens evaluated resulted genotoxic (Table 1). Cysplatin, 2-AAF, doxorubicin and carboplatin significantly increased SOSIF values with respect to control (untreated cells) in the presence of S9-mix thus indicating an indirect genotoxic effect of these compounds. However, cysplatin and carboplatin also significantly increased SOSIF values in the absence of exogenous S9-mix. Except for the case of cysplatin, the increasing in SOSIF values in E. coli PQ-37 cells treated with these mutagens was directly related with the increase of their concentration. In the absence of S9-mix metabolic activation, the lower concentration of cysplatin yielded the highest SOSIF value probably due to its cytotoxic effect at higher concentrations.
On the other hand, the extracts of P. orbicularis and P. caribaea did not increase the SOSIF values. There wasn’t a correlation (r2 = 0.23; r2 = 0.01 respectively, p < 0.05) between the extract concentrations and SOSIF values obtained, indicating that these plants extracts were not genotoxic to E. coli PQ-37 cells. In contrast, the C. citratus aqueous extract was genotoxic in E. coli PQ-37 cells starting from the concentrations of 2 mg/mL (r2 = 0.95 p < 0.05). This behavior was observed in both the presence and the absence of exogenous S9-mix (Table 2).
DISCUSSION
The modifications to the original SOS Chromotest procedure [2] were addressed to switch from colorimetric to fluorescent substrates to facilitate the analysis of pigmented samples. Furthermore, the work was aimed at adapting the assay to a microanalytical scale, thus reducing the final assay volume and automatizing readings. We evaluated the β-galactosidase activity and its substrate concentration in the assay by using a physical mutagen (γ rays) as inducer of DNA damage and triggerer of the SOS response [12-14]. The SOSIF values obtained using the selected substrate concentration and reaction time resulted quite similar to those obtained with the use of the colorimetric variant [2]. However, this substrate concentration was 10 times lower than that of the ONPG concentration used in the original assay along with the fact that the total reaction volume was 20-fold reduced in the modified protocol, which reduces reagents costs in 17%. Also, semiautomatic readings allow for reducing data acquisition time from one hour to approximately 5 minutes. Such advantages are relevant with regard to the rapid evaluation of a higher number of small-volume samples and to potential costs reduction of the test.
Additionally, a characterization of the modified procedure presented here was performed by calculation of kinetical parameters. On this matter, it is worth to mention that this is an ‘ex vivo’ assay that uses small volumes of cell lysates, which impossible to determine the active enzyme concentration and therefore, kcat calculation. For this reason, we decided to determine an apparent Vmax and Km from Lineweaver-Burk plots in order to compare it with the Km reported for other β-galactosidase assays. The Km obtained (1.9 mM) in the presence of 4-methylumbelliferyl galactopyranoside was similar to the lower values reported in other microorganisms using the ONPG colorimetric substrate from the unmodified SOS Chromotest. Km values of 1.5, 1.8 and 2.6 mM were found for Kluyveromyces lactis, Penicillium chrysogenum, Bifidobacterium infantis and Bacillus sp., respectively [18].
The modified protocol used in the present study was effective to detect genotoxicity of several known mutagens. Mutagen selection was performed taken into account the criteria to include DNA damage inducers that follow different mechanisms. For instance, bleomycin induce simple and double strand breaks, cysplatin and carboplatin cause DNA crosslinks, 2-acethyl-aminofluorene (2-AAF) forms DNA adducts, while mitomycin C is an alquil agent and doxorubycin is an intercalant agent [19]. These mutagens induced SOS response, because they directly or indirectly (by damage processing) were able to generate single strand breaks, which is the proposed signal to trigger emergency response. They were previously reported as SOS inducers [20-22].
The usefulness of the proposed procedure was also proved through the evaluation of pigmented natural extracts. The three plants extracts were selected because of their properties and prospective use in human health related issues. P. orbicularis extract has antiviral properties, P. caribaea has antioxidant activity [23] and C. citratus is one of the natural extracts most used by the Cuban population due to some of its pharmacological properties [24].
The results of the genotoxicological evaluation of P. orbicularis extracts obtained in our fluorescent SOS test version are in agreement with those reported previously in 2002, using the Umutest and Ames test. Conversely, it has been also reported that this extract could be genotoxic in other biological models although this positive genotoxicity was associated to its cytotoxicity in vitro and in vivo. In addition, the P. caribaea extract did not induce primary DNA damage in E. coli PQ-37 cells. Negative toxicity of P. caribaea aqueous extract as well as of condensed tannins, the principal component of this extract, was already observed using SOS Chromotest models, respectively [15]. However, there is some evidence showing that tannins and other polyphenols compounds can be mutagenic. The mutagenicity of tannins greatly depends on their structural characteristics. For example, structure-function studies on compounds related to tannic and galic acids showed the latter as more mutagenic due to the presence of structural digaloil and free hydroxyl groups [15]. In contrast to the results obtained using P. orbicularis and P. caribaea extracts, the C. citratus aqueous extract induced primary DNA damage starting from the 2 mg/mL concentration, indicating its genotoxicity in the model studied. We have also observed on agarose gel electrophoresis (data not shown) that this plant extract induces single and double DNA strand breaks on pUC18 plasmid, perhaps generating an oxidative-based mechanism. Studies related to C. citratus genotoxicity are contradictory [14].
In conclusion, the modified version proposed here was sensible to known chemical and physical mutagens. SOSIF values obtained were high enough to be useful as positive controls in anti-genotoxicological studies. It was also advantageous to evaluate the genotoxicity of pigmented samples, like the plants extracts and mutagens such as mitomicin c and doxorubicin. It was less laborious than the use of blanks for each concentration, and allowed the analysis of very small volumes. To work at micronalytical scale is rapid, economic and represents a valid alternative for the screening of a large variety of samples.
ACKNOWLEDGMENTS
The authors wish to thank Gisela Barrera, Mercedes Guerra and Sonia Altanés for technical assistance during irradiation of cells. This work was supported by the PRN/7-1/3-2002 project of the Nuclear Agency of the Cuban Ministry of Science, Technology and Environment (CITMA).
REFERENCES
1. Ames BN, Durston WE, Yamasaki E, Lee FD. Carcinogens are mutagens: a simple test system combining liver homogenates for activation and bacteria for detection. Proc Natl Acad Sci USA. 1973; 70(8):2281-5.
2. Quillardet P, Huisman O, D’Ari R, Hofnung M. SOS chromotest, a direct assay of induction of an SOS function in Escherichia coli K-12 to measure genotoxicity. Proc Natl Acad Sci USA. 1982;79(19):5971-5.
3. Quillardet P, Hofnung M. The SOS chromotest: a review. Mutat Res. 1993; 297(3):235-79.
4. Zounkova R, Odraska P, Dolezalova L, Hilscherova K, Marsalek B, Blaha L. Ecotoxicity and genotoxicity assessment of cytostatic pharmaceuticals. Environ Toxicol Chem. 2007;26(10):2208-14.
5. Leet JE, Liu X, Drexler DM, Cantone JL, Huang S, Mamber SW, et al. Cytotoxic xanthones from Psorospermum molluscum from the Madagascar rain forest. J Nat Prod. 2008;71(3):460-3.
6. Westerink WM, Stevenson JC, Lauwers A, Griffioen G, Horbach GJ, Schoonen WG. Evaluation of the Vitotox and RadarScreen assays for the rapid assessment of genotoxicity in the early research phase of drug development. Mutat Res. 2009;676(1-2):113-30.
7. Vidal LS, Alves AM, Kuster RM, Lage C, Leitao AC. Genotoxicity and mutagenicity of Echinodorus macrophyllus (chapeu-de-couro) extracts. Genet Mol Biol. 2010;33(3):549-57.
8. Vicuña GC, Stashenko EE, Fuentes JL. Chemical composition of the Lippia origanoides essential oils and their antigenotoxicity against bleomycin-induced DNA damage. Fitoterapia. 2010;81(5):343-49.
9. Afanassiev V, Sefton M, Anantachaiyong T, Barker G, Walmsley R, Wolfl S. Application of yeast cells transformed with GFP expression constructs containing the RAD54 or RNR2 promoter as a test for the genotoxic potential of chemical substances. Mutat Res. 2000;464(2):297-308.
10. Ichikawa K, Eki T. A novel yeast-based reporter assay system for the sensitive detection of genotoxic agents mediated by a DNA damage-inducible LexA-GAL4 protein. J Biochem. 2006;139(1):105-12.
11. Podgorska B, Wegrzyn G. A modified Vibrio harveyi mutagenicity assay based on bioluminescence induction. Lett Appl Microbiol. 2006;42(6):578-82.
12. Quillardet P, Frelat G, Nguyen VD, Hofnung M. Detection of ionizing radiations with the SOS Chromotest, a bacterial short-term test for genotoxic agents. Mutat Res. 1989;216(5):251-7.
13. Kozubek S, Ogievetskaya MM, Krasavin EA, Drasil V, Soska J. Investigation of the SOS response of Escherichia coli after gamma-irradiation by means of the SOS chromotest. Mutat Res. 1990;230(1):1-7.
14. Fuentes JL, Alonso A, Cuetara E, Vernhe M, Alvarez N, Sanchez-Lamar A, et al. Usefulness of the SOS Chromotest in the study of medicinal plants as radioprotectors. Int J Radiat Biol. 2006;82(5):323-9.
15. Prieto E, Cañet F. Aspectos a considerar en el dosímetro Fricke. Tecnol Quím. 1990;2:19-20.
16. Salvo NJ, Castillo A, Fernández A, Bouzó L, Torres K, González F. Optimización de la producción del anticuerpo monoclonal iorT3 en biorreactores de fibra hueca. Biotecnol Apl.1994;11(2):160-4.
17. Kevekordes S, Mersch-Sundermann V, Burghaus CM, Spielberger J, SchmeiserHH, Arlt VM, et al. SOS induction of selected naturally occurring substances in Escherichia coli (SOS chromotest). Mutat Res. 1999;445(1):81-91.
18. BRENDA. BRaunschweig ENzyme DAtabase. Braunschweig: Institute of Biochemistry and Bioinformatics at the Technical University of Braunschweig. c2011 - [cited 2011 Dec 15]. Available from: http://www.brenda-enzymes.org
19. Takimoto CH, Ng CM. Pharmacokinetics and Pharmacodynamics. In: DeVita VT, Lawrence TS, Rosenberg SA. Cancer: Principles and Practice of Oncology. 8th edition. Philadelphia: Lippincott, Williams & Wilkins; 2008. p. 392-401.
20. Ohta T, Nakamura N, Moriya M, Shirasu Y, Kada T. The SOS-function-inducing activity of chemical mutagens in Escherichia coli. Mutat Res. 1984;131(3-4):101-9.
21. Quillardet P, de Bellecombe C, HofnungM. The SOS Chromotest, a colorimetric bacterial assay for genotoxins: validation study with 83 compounds. Mutat Res. 1985;147(3):79-95.
22. Overbeck TL, Knight JM, Beck DJ. A comparison of the genotoxic effects of carboplatin and cisplatin in Escherichia coli. Mutat Res. 1996;362(3):249-59.
23. Fuentes JL, Vernhe M, Cuetara EB, Sánchez-Lamar A, Santana JL, Llagostera M. Tannins from barks of Pinus caribaea protect Escherichia coli cells against DNA damage induced by γ-rays. Fitoterapia. 2006;77(2):116-20.
24. Alonso Martín A, Almeida Varela E. Las plantas como radioprotectores potenciales frente a la radiación ionizante. Nucleus. 2008;(44):3-7.
Received in September, 2011.
Accepted for publication in February, 2012.
Elizabeth B Cuétara. Instituto Nacional de Oncología y Radiobiología. Esquina 29 y F, Vedado, Plaza, La Habana, Cuba. E-mail: ecuetara@infomed.sld.cu.














