INTRODUCTION
Equine influenza is a highly contagious respiratory disease in horses caused by influenza A viruses. Equine influenza virus belongs to the Orthomyxoviridae family and there are two different subtypes, H7N7 (formerly subtype 1) (1) and H3N8 (formerly subtype 2) (2). Characteristically, influenza spreads rapidly in a susceptible population. There have been very few reports worldwide regarding with the subtype H7N7 virus in the last 30 years; however outbreaks caused by H3N8 viruses occur annually (3). It is of paramount importance to have specific and sensitive virus detection systems available for a rapid diagnosis of equine influenza. Traditionally, the gold standard has been virus isolation from nasopharyngeal swabs using embryonated hen eggs. Other diagnostic tests employed include virus culture in Madin Darby canine kidney (MDCK) cells or measuring an increase in antibody titer in paired sera by the hemagglutination inhibition assay (4). These established techniques are time consuming and laborious. Furthermore, the sensitivity of virus isolation depends on the presence of infectious particles, and some virus strains are difficult to isolate (5). Serology provides only retrospective data which do not facilitate prompt intervention and appropriate case management. In recent years, enzyme-linked immunosorbent assays such as Directigen FluA (Becton Dickson) and RT-PCR have been introduced in some laboratories to provide more timely results (6,7).
Nowadays, rRT-PCR assays are increasingly used for the detection of the equine influenza virus genome; however most of these tests are designed on the basis of probes, which are expensives and very difficult to acquire for our country (8-11).
A useful alternative has been the SYBR Green-I based real-time assay, which has proven to be one of the most effective tools in the rapid and differential detection of a variety of viral pathogens (12-19). The Light Cycler PCR system from Roche Molecular Biochemicals allows the high-speed thermal cycling and quantitative RT-PCR (qRT-PCR) using SYBR Green-I, which is a cost efficient option.
In Cuba, equine influenza is an exotic disease. That is why it is very useful to have an rRT-PCR system suitable for detection of this viral agent at the Virology Laboratory at CENSA.
The purpose of this study was to standardize an rRT-PCR assay, based on SYBR Green-I coupled with melting curve analysis for the detection of equine influenza virus (gene M) and the typing of hemagglutinin gene.
MATERIALS AND METHODS
For rRT-PCR assay standardization, an RNA of equine influenza virus, gently donated by the Central Veterinary Laboratory of Algete Spain, was used as positive control.
The synthesis of cDNA was performed using random priming and M-MLV reverse transcriptase (Promega, Madison, WI, USA), as previously described by Díaz de Arce et al. (20).
The primers M+25 (5’-AGATGAGTCTTCTAACCGAGGTCG-3’) and M-124 (5’-TGCAAAAACATC TTCAAGTCTCTG-3’), designed by Spackman, 2002, were used to amplify a 99 bp amplicon from a conserved region in all type A influenza virus matrix genes (21).
A second set of primers was used for subtyping the hemagglutinin (HA) gene. A 373 bp region of this gene (nucleotides 566 to 938) was amplified using primer HA3F (5’-GAATGTGACAATGCCTAAC-3’) and primer HA3R (5’-GATGCTTCCATTTGGTGTA-3’).
The rRT-PCR to detect the matrix gene was optimized and carried out using the SYBR Green Quantitative RT-PCR Kit (Roche Diagnostics, Manheim, Germany). Different concentrations of primers (0.3, 0.4, 0.5 μM) and MgCl2 (2.0, 3.0 y 4.0 mM) were evaluated.
All rRT-PCR reactions were performed in duplicate and conducted on the LightCycler 1.5 (Roche Diagnostics, Manheim, Germany) instrument. The following protocol was used: 10 min at 95°C, followed by 45 cycles at 95°C for 10 s, 55°C for 10 s and 72°C for 20 s. After PCR cycles, a DNA melting curve was generated (0 s at 95°C, 15 s at 65°C, with a ramping time of 20°C/s and 0 s at 95°C with a ramping time of 0.3°C/s), in order to discriminate between specific and nonspecific amplification products. In this study, the assay detection limit was determined by the efficacy of the entire method, including the sensitivity of the primers and the optimization of the procedure.
To determine the assay detection limit in terms of RNA copy numbers, an in vitro-transcribed RNA of the M gene targeted region was obtained. After reverse transcription, PCR was performed with the reverse primer coupled with a T7 promoter. The amplified products were checked by 2% agarose gel electrophoresis, stained with ethidium bromide (0.5 g/mL) and purified using Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, E.U.A.), according to the manufacturer’s recommendations. The RT-PCR purified products were directly in vitro transcribed from the T7 polymerase promoter site using a MEGAscript® Kit (Ambion, Carlsbad, CA, E.U.A.), following the manufacturer’s recommendations. Briefly, the transcription reaction was performed using an RNA Polymerase Enzyme Mix, and a TURBO DNase treatment was carried out to remove the template DNA. Afterwards, the MEGAclear™ Kit was employed specifically for purifying RNA. Finally, the RNA transcript was quantified by spectrophotometry. Thus, a total of 4.5x1013 RNA copies/µl was obtained. Also, standard curves based on gene copy different numbers were generated, and reaction efficiencies were calculated by the LightCycler software (Version 4.05).
The assay detection limit in terms of RNA copy numbers was determined by testing sequential ten-fold dilutions of the in vitro transcribed RNA in nuclease-free water (Promega, Madison, USA). The assay specificity was tested on the RNA of equine viral arteritis, equine herpesvirus-1 and equine herpesvirus-4. Each strain used was tested in triplicate.
A second independent rRT-PCR protocol was carried out with HA gene primers. rRT-PCR conditions used with the second set of primers were the same as those used with the matrix primers.
RESULTS AND DISCUSSION
Finally, 3.0 mM and 0.4μM concentrations of MgCl2 and primers were established and used in further experiments. The final PCR mixture contained 0.8 μl each of forward and reverse primers (final concentration of each, 0.4 μM), 2 μl of Fast Start DNA Master SYBR Green-I, 1.6 μl of MgCl2 (final concentration 3.0 mM), 5 μl of template, and made up to 15 μl with nuclease free water. The optimal alignment temperature was 55oC (Figure 1 and 2).

Figure 1 Evaluation of different MgCl2 concentrations./Evaluación de diferentes concentraciones de MgCl 2. (A) Amplification curves obtained from RNA equine influenza virus using different concentrations of MgCl2. (B) Analysis of melting curves showing that the optimal concentration of MgCl2 is 3 mM. Nuclease-free water was used as negative amplification control./ (A) Curvas de amplificación obtenidas a partir de ARN del virus de influenza equina usando diferentes concentraciones de MgCl 2 . (B) El análisis de curvas de disociación mostró que la concentración óptima de MgCl 2 es 3 mM. Se usó agua libre de nucleasas como control de amplificación.
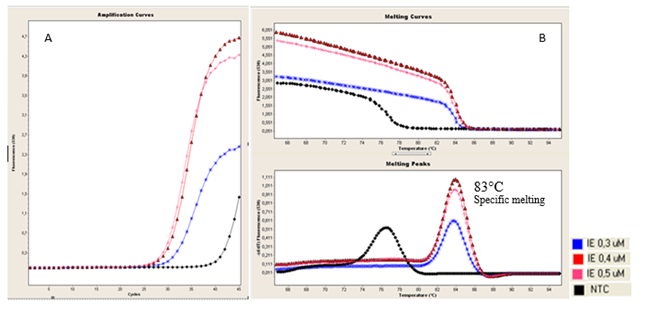
Figure 2 Evaluation of different primer concentrations./Evaluación de diferentes concentraciones de cebadores. (A). Amplification curves obtained from RNA equine influenza virus using different concentrations of primers. (B). Analysis of melting curves showing that the optimal concentration of primers is 0.4 µM. Nuclease-free water was used as negative amplification control. / (A). Curvas de amplificación obtenidas a partir de ARN del virus de influenza equina usando diferentes concentraciones de cebadores . (B). El análisis de curvas de disociación mostró que la concentración óptima de cebadores es 0.4 µM. Se usó agua libre de nucleasas como control de amplificación.
In terms of the matrix gene copy number, the detection limit was 4.5 gene copies/μl of in vitro transcribed RNA.
To determine the linearity of the reaction and the PCR efficiency, the threshold cycle values of individual dilutions were plotted against the gene copy number. The linear range obtained for the transcript in the assay generated a typical standard curve from 107 until 100 gene copies/µl in terms of RNA copy number (Figure 3 A). The correlation coefficient (R2) was 0.99 for the RNA gene copies detected in nuclease free water. The error of standard curve was 0.0139, which is in the acceptance range for this type of test (less or equal to 0.02) (Figure 3 B).

Figure 3 Amplification by SYBR Green-I based rRT-PCR assay for the detection of equine influenza virus. (A) The curve shows amplification of eight serial dilutions (Log 10) of the in vitro gene transcript M. ( B ) The standard curve shows the linear range of the assay considering the number of detectable RNA transcribed copies./ Amplificación por rRT-PCR basado en SYBR Green-I para la detección del virus de influenza equina. (A) La curva muestra la amplificación de ocho diluciones seriadas (Log 10) del transcripto in vitro del gen M. (B) La curva estándar muestra el rango lineal del ensayo considerando el número de copias detectable del ARN transcripto.
The rRT-PCR assay showed to be specific for the equine influenza virus. Amplification specificity was carried out by the melting curve analysis of the amplified products in rRT-PCR assay. The specific amplifications of equine influenza virus were also confirmed by the detection of a band of 99 bp by agarose gel electrophoresis (data not shown). No specific amplification curves were obtained with any of the other viruses tested such as equine viral arteritis, equine herpesvirus-1 and equine herpesvirus-4 (Figure 4).

Figure 4 Evaluation of analytical specificity. (A) Amplification curves obtained from RNA of equine influenza virus, equine viral arteritis, equine herpesvirus-1 and equine herpesvirus-4). (B) Melting curve analysis showed the rRT-PCR to be specific for equine influenza virus but not for the other viruses tested./ Evaluación de la especificidad analítica. (A) Curvas de amplificación obtenidas a partir de ARN del virus de influenza equina, arteritis viral equina, herpesvirus equino-1 y herpesvirus equino-4. (B) Análisis de curvas de disociación mostró que el rRT-PCR es específico para el virus de influenza equina y no así para los otros virus evaluados.
The SYBR Green-I based real time RT-PCR assay showed specific amplification and melting curves when the second set of primers against HA gene was evaluated (Figure 5).
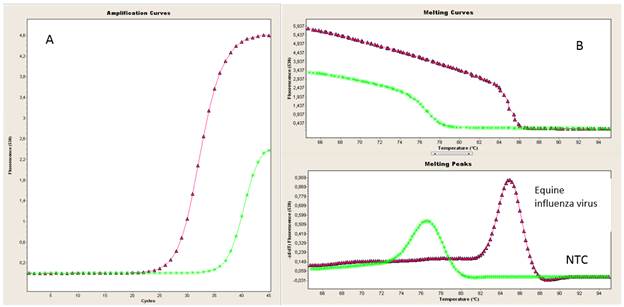
Figure 5 Evaluation of rRT-PCR using HA gene primers. Amplification curves (A) and melting curves (B) obtained from RNA equine influenza using HA3F and HA3R primers. Nuclease-free water was used as negative amplification control./ Evaluación de rRT-PCR utilizando los cebadores del gen para HA. Curvas de amplificación (A) y de disociación (B) obtenidas a partir de ARN de influenza equina usando los cebadores HA3F y HA3R. Se usó agua libre de nucleasas como control de amplificación.
The major challenge faced by equine veterinarians when confronted with a case of respiratory tract infection is to determine the contagious nature of the disease in order to properly manage the affected animal and to reduce the risk of exposure to other horses. This requires the availability of fast and reliable diagnostic tests.
The real-time technology provides highly sensitive, specific as well as rapid detection and quantification of viral genomes and should be carried out with a low contamination risk at a reasonable cost (22,23). Nowadays, RT-PCR in real-time assays is increasingly used for the detection of equine influenza virus in nasal secretions (24). Different assays based on this tecnology using TaqMan probes have been documented in the OIE Manual (8,9). Nevertheless, the rRT-PCR assays based on SYBR Green are potentially less influenced by mismatches because they are not influenced by sequence variation in the probe target region; they have relative simplicity and the cost of SYBR Green-I is lower compared to probes (25).
The proposed SYBR Green-I based real-time RT-PCR assay coupled with melting curve analysis provides a reliable, fast, sensitive, and specific method for the detection of equine influenza virus, althought it must be evaluated in clinical samples. It can be used in the case of a possible emergency of this agent in Cuba.