Introduction
Nowadays, 292 million people are chronically infected with hepatitis B virus (HBV), as a result, 1.4 million of people die each year in the world.1,2) This infectious disease is in second place after tuberculosis in terms of mortality and the number of infected people with hepatitis is higher than the number of infected people with HIV. However, over 80% of people infected with hepatitis do not have access to prevention, testing, and treatment.3)
Currently, the HBV virus vaccine, which is a recombinant hepatitis B surface antigen protein based on the HBV-S region, is used to the prevention of this disease in the world. This protein (consists of 226 amino acids) contains only one immunogenic determinant "a". It is common to all genotypes of HBV and cannot provide complete protection against the virus.4)
It is known that proteins encoded by PreS1 (108 a.a.), PreS2 (55 a.a.), and S (226 a.o.) regions of the virus genome are considered highly immunogenic and a part of its outer surface. It is considered that the PreS2-S (PreS2 + S) region of the HBV surface protein (containing 281 amino acids) has polyalbumin-binding activity, which indicates the complementarity of hepatocyte surface receptors.
Recent studies have shown that the presence of PreS2-S protein in the hepatitis B virus vaccine contributes to a significant increase in the immunogenicity of the vaccine.5,6,7,8,9 Today, various expression systems are used to obtain this protein, including yeast expression system.10,11
Pichia pastoris methylotrophic yeast is one of the widely used microorganisms for the production of heterologous proteins. The advantages of the P. pastoris expression system are: the accumulation of significant biomass during cultivation on inexpensive nutrient media, the absence of endotoxins and pyrogens, a higher level of synthesis of recombinant proteins and the ability to synthesize recombinant protein in a nutrient medium.12,13,14,15)
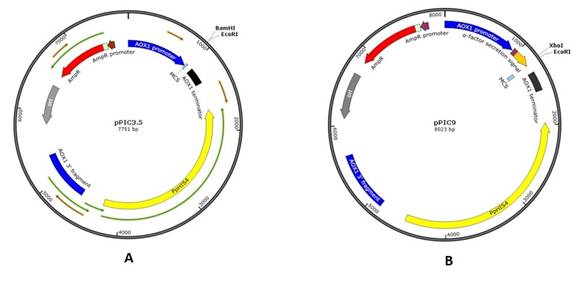
Various vectors are currently available for use in this expression system. In the present work, the transfer vectors pPIC3.5 (7751 b.p) and pPIC9 (8023 b.p) containing the nucleotide sequences of the P. pastoris genome, including the AOX1 promoter, HIS4 were used in order to obtain new recombinant plasmids encoding the PreS2-S region of the HBV of genotype D commonly widespread in Uzbekistan.16
Materials and Methods
In the experiments, enzymes and reagents from Thermo Fisher Scientific (USA), New England Biolabs (USA), Sigma-Aldrich (Merck, Germany), SBS Genetech (China), Panreac (Germany), Biosset (Russia), Himedia (India) were used.
Preparation of vectors and insertion (gene) for ligation
A bacterial strain of Escherichia coli containing the transfer vectors pPIC3.5 and pPIC9 (Invitrogen, Cat. no. K1710-01) was cultivated on Luria Bertani medium in the presence of ampicillin (30 μg/mL) at a temperature of 37°C.17,18)
One microgram each of vectors pPIC3.5 and pPIC9 was digested sequentially with BamH I/EcoR I and Xho I/EcoR I, respectively.18 Linear DNA molecules with length of 7745 bp and 7999 bp were isolated after electrophoresis in a 0.7% agarose gel by the method of electroelution.
The PreS2-S gene was prepared from the plasmid pBacPAK-8-Polh-PreS2-S by digestion with restriction enzymes BamHI/EcoRI for insertion into the transfer vector pPIC3.5 and XhoI/EcoRI for pPIC9.16,19 The nucleotide sequence of Pres2-S gene fragment can be found in ENA with accession number LR745788.20
Ligation and transformation of the obtained plasmids into electrocompetent cells of E. coli Neb-5α. Analysis of the obtained clones
Ligation was performed using a T4 phage DNA-ligase with a prepared vector into a molar ratio of 1:10 (vector: insert) according to the standard method.17) Then, the ligase mixture was transformed into competent cells of the E. coli NEB-5α.
The size of DNA plasmid isolated from clones that were obtained after transformation with a ligase mixture was determined by the method of agarose gel electrophoresis. Clones containing DNA with a molecular weight of 8.707 and 8.980 kb were investigated by PCR analysis using the following specific primers:
For pPIC3.5-PreS2-S
1. 5`-CGGATCCAAAAATGTCTCAGTGGAAC-3`
2. 5`-TGTTGAATTCAATGTATACCCAAA-3`
For pPIC9-PreS2-S
1. 5`-CCTCGAGAAAAATGTCTCAGTGGAAC-3`
2. 5`-TGTTGAATTCAATGTATACCCAAA-3`
After that, the products of amplification were analyzed by electrophoresis in a 1% agarose gel. DNA of the recombinant plasmids identified by PCR was also confirmed with restriction analysis. For this, a comparative analysis of the recombinant clones and the initial vector plasmids pPIC3,5 and pPIC9 were restricted by the EcoRI site.18) The molecular size of the obtained fragments was determined by electrophoresis in a 0.7% agarose gel; 1 kb Plus DNA Ladder from Invitrogen (Cat. No 10787018) was used as a molecular weight marker.
Transformation of the obtained recombinant plasmids to the P. pastoris yeast strain and identification of the obtained clones
The transformation of the obtained recombinant plasmids containing the target genes to the P. pastoris was conducted by the method of electroporation.18) Integration of the target gene into the genome of GS115 P. pastoris strain was detected by PCR using the following primers:
ELISA and Western blotting analysis of recombinant PreS2-S protein
ELISA was performed according to Clark and Adams.21 The recombinant PreS2-S protein was determined using test systems of the “ORTO Diagnostics” (USA) and “Diagnostic test systems of ICPS AS RUz” (Uzbekistan). The recombinant protein was detected by the corresponding antibody conjugated with alkaline phosphate diluted in conjugate buffer (PBS-TPO, pH 7.4) according to the suppliers’ specifications. Absorbance values were measured at 450 nm with a microplate reader (ELx800 Universal Microplate Reader, Bio-Tek Instruments Belgium). A sample was considered to be positive if its absorbance value was greater at least two times than absorbance values of the negative control. Whole cell-lysate of non-containing PreS2-S gene of GS115 P. pastoris cells was used as a negative control. For the analysis of Western blotting was used a commercial kit “Western Breeze, Chromogenic Western Blot Immunodetection Kit” (Invitrogen, Cat. WB7105).
Results and Discussion
According to their physical maps (Fig. 1), plasmids pPIC3.5 (A) and pPIC9 (B) were processed sequentially with restriction enzymes BamH I/EcoR I and Xho I/EcoR I in order to clone new inserts. The ligation of foreign DNA encoding the amino acid sequence of the protein into these sites, places the heterologous gene under the control of the promoter of the AOX1 gene.18)
BamHI:
5′-G↓GATCC-3′ (pPIC3.5)
3′-CСTAG↓G-5′
XhoI:
5′-C↓TCGAG (pPIC9)
3′-GAGCT↓С-5′
EcoRI:
5′-G↓AATTC-3′ (pPIC3.5 and pPIC9)
3′-CTTAA↓G-5′
As the results, fragments of the vectors pPIC3.5 and pPIC9 with “sticky” ends that allowing for “directed” ligation were obtained. Linear DNA plasmids with a length of 7745 bp and 7999 bp were isolated after agarose gel electrophoresis. They were processed with alkaline phosphatase for avoid the "closure" of the vectors.
The PreS2-S gene was isolated from plasmid pBacPAK-8-Polh-PreS2-S by digestion with appropriate restriction enzymes BamHI/EcoRI and XhoI/EcoRI.16,19
The digested transfer vectors were ligated to the fragment containing PreS2-S cDNA of the HBV region in a molar ratio of 1:10 by using T4 phage DNA ligase. After that, the obtained ligase mixture was transformed into the competent cells of E. coli NEB-5(. The identification of the formed clones was performed by PCR using the primers for the PreS2-S region. Then, PCR products were analyzed by electrophoresis in 1% agarose gel (Fig. 2).
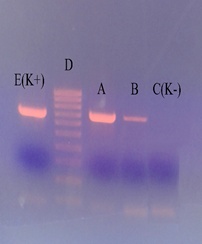 A- pPIC3.5-PreS2-S; B - pPIC9-PreS2-S; C (K-) - DNA isolated from the blood of HBV negative people; D - DNA line (Marker); E (K +) - DNA isolated from the blood of HBV "positive" patients.
A- pPIC3.5-PreS2-S; B - pPIC9-PreS2-S; C (K-) - DNA isolated from the blood of HBV negative people; D - DNA line (Marker); E (K +) - DNA isolated from the blood of HBV "positive" patients.Fig. 2. Gel electrophoresis of transformants containing the target gene.
The presence of a DNA fragment (A and B) with a molecular size (956 bp) corresponding to the mass of the DNA fragment amplified from the HBV positive sample (E) indicated the presence of an insert in the studied plasmids. Thus, clones containing the PreS2-S region of HBV DNA were detected by PCR. Comparative restriction analysis of recombinant plasmids pPIC3.5-PreS2-S and pPIC9-PreS2-S with the original vectors pPIC3.5, pPIC9 by the EcoR I single site indicated that the molecular weight of recombinant plasmids corresponds to theoretical calculations on physical maps of these plasmids by the results of electrophoresis in a 0.7% agarose gel (Fig. 3). The linear forms of the recombinant plasmids pPIC3.5-Pres2-S and pPIC9-Pres2-S corresponding to 8707 bp and 8980 bp by the digestion with the EcoR I restriction enzyme were obtained.
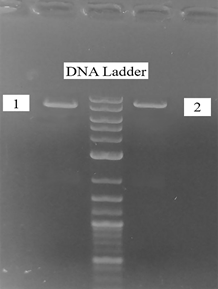
Fig. 3 Restriction analysis of pPIC3.5-PreS2-S (EcoR I/8707 bp). 1- pPIC3.5/EcoR I (7751 bp); 2- pPIC3.5-PreS2-S/EcoR I (8707 bp)
Then, the transformation of recombinant plasmids pPIC3.5-PreS2-S and pPIC9- PreS2-S linearized in the HIS4 region by Sal I nuclease into the strain of P. pastoris yeast was conducted. Transformants (His +) were selected in histidine-deficient media. Obtained recombinant clones and integration of the target gene into the P. pastoris genome were identified by PCR using the α-factor primer (for pPIC9 only) and the 5´ AOX1 primer paired with the 3´ AOX1 primer.18
Several selected colonies of transformants were cultivated to the stage of the exponential phase (OD=2 at 600 nm) in a medium containing glycerin. The expression was induced by the addition of methanol until a final concentration of 0.5%, over 120 h of cultivation. The level of synthesis of recombinant protein and specificity were determined by the method of solid-phase enzyme linked immunoassay analyses (ELISA). For this purpose the samples (whole-cell lysates) were taken serially at 24 h intervals. Hepatitis-B Vaccine (rDNA) 20 µg/mL, Serum Institute of India and Euvax-B (Hepatitis B Vaccine [rDNA]), Sanofi Pasteur Korea Ltd., were used as the calibration standard (Table 1).
Table 1 Determination of recombinant Pres2-S protein level in whole-cell lysates of P. pastoris by ELISA depending on cultivation time.
| The cultivation time after induction with methanol, hours | Result of ELISA (OD value at 450 nm) | The amount of PreS2-S protein in the test sample |
|---|---|---|
| 24 | 0.612 | ≈ 5 ng/mL |
| 48 | 1.100 | ≈ 10 ng/mL |
| 72 | 1.998 | ≈ 20 ng/mL |
| 96 | >3.000 | ≥ 26.5 ng/mL |
| 120 | 2.890 | ≈ 25 ng/mL |
| Euvax-B (Hepatitis B Vaccine [rDNA]) 20 µg/mL diluted: | ||
| 1:4,000 | 0.532 | ≈ 5 ng/mL |
| 1:2,000 | 0.971 | ≈ 10 ng/mL |
| 1:1,000 | 2.178 | ≈ 20 ng/mL |
| 1:750 | >3.000 | ≥ 26.5 ng/mL |
According to the results of ELISA, the optimal cultivation time was 96 h after induction with methanol.
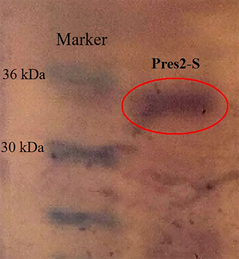
The results of electrophoresis in PAGE and immunoblotting showed correspondence of the obtained recombinant protein to the expected molecular weight (about 34 kDa) and high specificity to the HBV antibodies (Fig. 4).
Conclusion
New recombinant plasmids pPIC3.5- PreS2-S and pPIC9- PreS2-S containing the cDNA PreS2-S region of the HBV surface antigen were cloned and recombinant strains of P. pastoris producing the target protein were obtained on their basis. The study of immunogenicity (humoral and cellular immunity) is required and may provide conclusive evidence of that the PreS2-S protein is an ideal candidate for a new generation of hepatitis B vaccines.