Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista de Salud Animal
versión impresa ISSN 0253-570X
Rev Salud Anim. v.32 n.1 La Habana ene.-abr. 2010
Short communication
MOLECULAR CHARACTERIZATION OF A Herpesvirus bovino 1 CUBAN STRAIN
CARACTERIZACIÓN MOLECULAR DE UNA CEPA CUBANA DE Herpesvirus bovino 1
Majela Rodríguez Medina*, Maritza Barrera Valle*, Monika Engels**, M. Ackermann**
*Centro Nacional de Sanidad Agropecuaria (CENSA). Autopista Nacional y Carretera de Jamaica, Apartado 10, San José de las Lajas, La Habana, Cuba. E-mail: majela@censa.edu.cu; **Institute of Virology, University of Zürich. Winterthurerstrasse 266a, Zürich, Switzerland
ABSTRACT
In Cuba, infections caused by Bovine herpesvirus 1 (BoHV-1) are endemic; however clinical signs are moderate, possibly because the BoHV-1 circulating genotypes are not associated to severe infections but these are not known. In the present study a BoHV-1 strain isolated in Cuba was compared with BoHV-1 strains from other different countries. Viral DNA was analysed by adoption of a clustering system based on restriction enzyme analysis (REA) with HindIII, HpaI, PstI and SfiI, which led to identify the Cuban isolate as a BoHV-1.1.III strain. The Cuban BoHV-1 differed from all the reference strains analysed and displayed a restriction fragment pattern more similar to strains originated from the European continent. The inclusion of these results into the clustering database is a contribution for future studies in tracing back the origin of novel infectious bovine rhinotracheitis (IBR) outbreaks.
Key words: Bovine herpesvirus 1; infectious bovine rhinotracheitis; restriction enzyme analysis; clustering system
RESUMEN
En Cuba, las infecciones causadas por Herpesvirus bovino 1 (BoHV-1) son endémicas; sin embargo los signos clínicos son moderados, posiblemente porque los genotipos circulantes de BoHV-1 no están asociados con infecciones severas pero estos no se conocen. En el presente estudio se comparó un aislado de BoHV-1 proveniente de Cuba con cepas de BoHV-1 de diferentes países. El ADN viral fue analizado por adopción de un sistema de agrupamiento basado en análisis de restricción (REA) con las enzimas HindIII, HpaI, PstI y SfiI, el cual permitió identificar el aislado cubano como una cepa de BoHV-1.1.III. El BoHV-1 cubano difirió de todas las cepas de referencias analizadas y mostró un patrón de restricción similar a cepas provenientes del continente europeo. La introducción de estos resultados en la base de datos de agrupamiento es una contribución para futuros estudios relacionados con el origen de nuevos brotes de rinotraqueitis infecciosa bovina (RIB).
Palabras clave: Herpesvirus bovino 1; rinotraqueitis infecciosa bovina; análisis de restricción; sistema de agrupamiento
Bovine herpesvirus 1 (BoHV-1), classified as a member of the family Herpesviridae, subfamily Alphaherpesvirinae, genus Varicellovirus (1), is the causative agent of several infections from the respiratory and genital tracts of domestic cattle and other ruminant species. Based on Western Blot analysis of viral proteins with a panel of monoclonal antibodies, as well as on restriction endonuclease analysis of the viral nucleic acid and differential amplification by PCR (2, 3, 4), BoHV-1 strains are classified into two subtypes, BoHV-1.1 and BoHV-1.2, BoHV-1.2 being additionally grouping into BoHV-1.2a and BoHV-1.2b.
In Cuba, studying BoHV-1 and other cattle pathogens has been limited by economic problems. Nowadays, the interest in cattle production is increasing, and gaining profound knowledge on the epidemiological status of the BoHV-1 infection is getting importance. This implies the characterization of BoHV-1 strains that are circulating in the country. The purpose of the present study was to compare a BoHV-1 isolate from Cuba with BoHV-1 strains from other different countries using a clustering system based on restriction enzyme analysis.
Madin-Darby bovine kidney cells (MDBK; ATCC CCL 22) were grown in Eagle's minimal essential medium (Eagle's MEM, BioConcept's AMIMED, Switzerland) supplemented with 100 IU mL-1 penicillin, 100 mg mL-1 streptomycin and 7% foetal calf serum (FCS, Omnilab, Switzerland). In the maintenance medium, FCS was reduced to 2%. The cells were used for virus stocks production of the Cuban BoHV-1 strain E8, isolated in 1984 during an outbreak of keratoconjunctivitis (5) and BoHV-1 reference strains previously characterized by Wyss (6) (Table 1).
Virus purification and DNA extraction were conducted as previously described by Engels et al (7). DNA concentrations were determined by reading of absorbance at 260 nm (NanoDrop Technologies, Inc., Witec NanoDrop® ND-1000 Spectrophotometer®). The absorbance ratio 260/280 nm was always between 1.93 and 2.
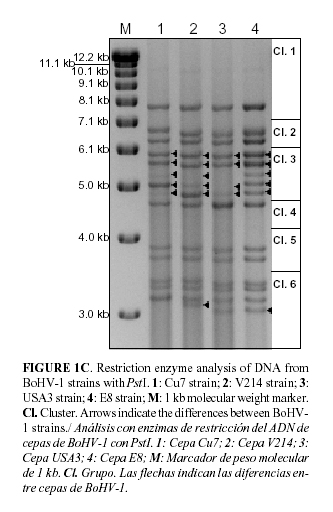
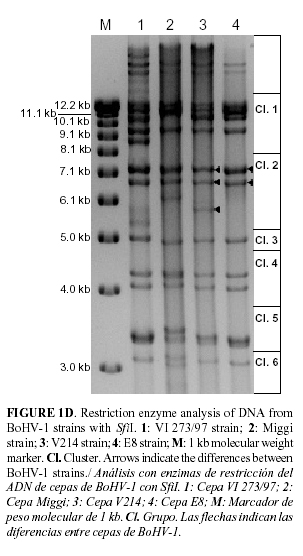
Aliquots containing 2 mg DNA were digested for 2 hr with HindIII, HpaI, PstI (Roche) and SfiI (BioLabs), respectively, under conditions recommended by the supplier in a final volume of 20 mL. The digestion products were separated by electrophoresis in 0.8% agarose gels containing 1 mg mL-1 ethidium bromide solution (BioRad). Gels were electrophoresed in TAE electrophoresis buffer (4 mM Tris-acetate, 1 mM EDTA; pH 8.0) between 18 and 24 hr at 60 V. For determinating the DNA fragment sizes, 1 Kb DNA Ladder (Gibco) with a size range of 500 bp to 12 kb and high molecular weight DNA markers (Invitrogen) with a size range of 9 kb to 48 kb were included. The bands were visualized and photographed using Quantity One® (Bio-Rad Laboratories, BioRad) and the molecular characterization was based on the clustering system by Wyss (6).
Comparative analysis of the resulting restriction enzyme patterns revealed that E8 HindIII pattern was more similar to one Canadian strain (BoHV-1 V214) and two European strains (BoHV-1 599/97 and BoHV-1 Cu7) (see Fig. 1A). As all those strains belonged to the BoHV-1.1 genotype, it was concluded that E8 belonged to the same genotype. Furthermore, isolate E8 was identified as BoHV-1.1.III strain, based on HindIII fragments E and F which appeared merging, and fragments C and D which showed higher molecular weight than the corresponding fragments of the other BoHV-1 strains.
Seven bands were obtained by E8 digestion with HpaI (see Fig. 1B). These results corresponded to the numeric code 421 based on the number of bands observed in clusters 1, 2 and 3, respectively.
E8 PstI pattern corresponded to the numeric code 125124. This code was also obtained from BoHV-1 V214, but differences in the molecular weight of the band 4 of cluster 6 were found (see Fig. 1C). Five fragments from E8 viral DNA were observed in cluster 3 and these were not observed from the American strain USA3 with 2 and 4 bands, respectively. Furthermore, bands 3, 4 and 5 of cluster 3 were observed at similar intensity for E8, and differences were found in BoHV-1 strains V214 and Cu7.
As it can be seen in Fig. 1D, the numeric code 421221 can be assigned to E8 SfiI pattern. It was more similar to the BoHV-1 strains V214 but only two bands in cluster 2 were obtained.
Restriction fragment pattern analysis has mostly been used for classification purposes knowing that the highest differences between BoHV-1.1 and BoHV-1.2 are restricted to distinct genomic regions characterized by loss or gain of restriction sites (8). It has also been used to differentiate between vaccine and field strains of BoHV-1 according to Hamelin et al (9). Moreover, according to systematic sequencing of individual isolates will be difficult to perform due to the high G + C content of BoHV-1 genomes (1), it is recommended the adoption of a clustering system based on restriction endonuclease analysis with enzymes HindIII, HpaI, PstI and SfiI in order to identifying the possible origins of BoHV-1 strains (6). This methodology was previously used to the identification of BoHV-1 strain origin, which had been isolated from imported semen and which had led to a new outbreak in 1983 in Switzerland (10).
The clustering data base includes about 100 different BoHV-1 strains previously analyzed by Wyss (6); in there, BoHV-1.1.III subtype corresponds with the 16% of BoHV-1.1 strains. It is important to note that all strains from Canada and U.S.A were 1.1.I subtype while 1.1.III subtype was only found in Europe. The highest number of strains with the same restriction fragment pattern (71%) was observed by digestion with HpaI. This can be explained by the small number of cleavage sites produced by this enzyme, which has been used for identification of the four major genotypes of BoHV-1 (11).
The main differences between E8 and the other bovine herpesviruses analyzed were found in the restriction fragment patterns obtained by digestion with PstI and SfiI. This is due to the high number of restriction sites for these enzymes that can be found in the genome of BoHV-1. The number of bands obtained in cluster 3 of E8 strain by digestion with PstI is not common and it was only found in strains from Canada, U.S.A, Holland and Belgium according to the table of classification performed by Wyss (2001). Taking into account the results obtained by Whetstone et al (12), it is possible to predict that the differences in E8 PstI pattern are associated with changes in the inverted repeat regions. These changes were observed in restriction endonuclease PstI digestion patterns of BHV-1.1 and BHV-1.2 isolates after one passage in host animal. Changes were also observed when virus was reactivated from latency or after superinfection with another strain of BHV-1 (13).
It was not possible to find a reference strain with the same four restriction fragment patterns as the BoHV-1 E8. The largest number of similar strains was originated from the European continent, but it has to be considered that comparison was made with strains mainly from this region. Considering that the animals and semen have been imported to the country only from Canada or USA, it is recommended to increase the number of entries from de American continent into the clustering data base. E8 DNA fragment pattern introduction in this database is a contribution for future studies in tracing back the origin of novel IBR outbreaks and is also a contribution to the updating of the Cuban surveillance epidemiological program but it is recommended to continue this study with other Cuban strains.
ACKNOWLEDGEMENTS
The authors thank A.E Metzler and M. Schwyzer for critical review and encouraging discussions. This research was supported by a grant from the Federal Commission for Scholarships for Foreign Students (FCS), Swiss Confederation.
REFERENCES
1. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds): Virus Taxonomy, VIIIth Report of the ICTV. 2005. Elsevier, Academic Press.
2. Rijsewijk FA, Kaashoek MJ, Langeveld JP, Meloen R, Judek J, Bienkowska-Szewczyk K, et al. Epitopes on glycoprotein C of bovine herpesvirus-1 (BHV-1) that allow differentiation between BHV-1.1 and BHV-1.2 strains. J Gen Virol. 1999; 80: 1477-1483.
3. D'Arce RCF, Almeida RS, Silva TC, Franco AC, Spilki F, Roehe PM, et al. Restriction endonuclease and monoclonal antibody analysis of Brazilian isolates of bovine herpesviruses types 1 and 5. Vet Microbiol. 2002; 88: 315-324.
4. Claus MP, Alfieri AF, Folgueras-Flatschart AV, Wosiacki SR, Médici KC, Alfieri AA. Rapid detection and differentiation of bovine herpesvirus 1 and 5 glycoprotein C gene in clinical specimens by multiplex-PCR. J Virol Methods. 2005; 128: 183-188.
5. Noda J, Nuñez A, García J. Aislamiento del virus de la rinotraqueitis infecciosa bovina en terneros con queratoconjutivitis. Rev Salud Anim. 1984; 6: 651-653.
6. Wyss SK. Etablierung und Anwendung einer auf der Restriktionsenzym Analyse basierenden Cluster-Technik und einer Datenbank des bovinen Herpesvirus Typ 1 für molekular-epidemiologische Untersuchungen. Veterinary Medicine Thesis, University of Zürich. 2001; 1-67.
7. Engels M, Gelderblom H, Darai G, Ludwing H. Goat Herpesviruses: Biological and physicochemical properties. J Gen Virol. 1983; 64: 2237-2247.
8. Engels M, Giuliani C, Wild P, Beck TM, Loepfe E, Wyler R. The genome of bovine herpesvirus 1 (BHV-1) strains exhibiting a neuropathogenic potential compared to known BHV-1 strains by restriction site mapping and cross-hybridation. Virus Res. 1986/87; 6: 57-73.
9. Hamelin C, Jacques C, Assaf R. Differentiation of vaccine and field strains of Bovine herpesvirus type 1 by restriction endonuclease analysis. Jpn J Vet Sci. 1990; 52(3): 461-467.
10.Ackermann M, Engels M. Pro and contra IBR-eradication. Vet Microbiol. 2006; 113: 293-302.
11.Edwards S, White H, Nixon P. A study of the predominant genotypes of bovid herpesvirus 1 found in the U.K. Vet Microbiol. 1990; 22: 213-223.
12.Whetstone C, Seal BS, Miller J. Variability occurs in the inverted repeat region of genomic DNA from bovine herpesvirus 1 respiratory, genital and bovine herpesvirus 5 encephalitic isolates. Vet Microbiol. 1993; 38: 181-189.
13.Whetstone C, Miller J, Bortner D, Van Der Maaten M. Changes in the restriction endonuclease patterns of four modified-live infectious bovine rhinotracheitis virus (IBRV) vaccines after one passage in host animal. Vaccine. 1989; 7: 527-532.
(Recibido 12-10-2009; Aceptado 20-12-2010)