Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista de Salud Animal
versión impresa ISSN 0253-570X
Rev Salud Anim. vol.35 no.1 La Habana ene.-abr. 2013
SHORT COMMUNICATION
Evaluation of simplified DNA extraction methods for Streptococcus suis typing
Evaluación de métodos simples de extracción de ADN para la tipificación de S. suis
Ivette Espinosa, M. Báez, María Irian Percedo, Siomara Martínez
Division of Molecular Biology, National Centre for Animal and Plant Health (CENSA), Apdo.10, San José de las Lajas, Mayabeque, Cuba. E-mail: espinosa@censa.edu.cu.
ABSTRACT
Streptococcus suis is a gram-positive bacterium that causes serious diseases in pigs and in humans with occupational risk. The DNA extraction methods for amplification of gene fragments by PCR for typing S.suis may be complex, and expensive chemical reagents and time consuming. The aim of this study was to evaluate a method for the rapid release of the genomic DNA from S.suis colonies by using a physical method based on heating and freezing; in this case, temperatures of 100oC and 95oC were tested. The results showed that DNA extraction directly from colonies by heating at 100oC could be useful for an easy genotyping of S.suis strains in a short time, while 95oC was not sufficient for DNA release. The detection limit of the PCR assay using DNA obtained by chemical purification was 0.5ng; considering the size of S.suis genome, it is possible to estimate that an adequate amount of cells are in a single S.suis colony to ensure the sensitivity of the PCR assay.
Key words: Streptococcus suis, direct colony PCR.
RESUMEN
Streptococcus suis es una bacteria grampositiva que causa serias enfermedades en cerdos y humanos con riesgo profesional. Los métodos de extracción de ADN para la amplificación de fragmentos de genes por PCR para la tipificación de S.suis pueden resultar complejos, consumir reactivos costosos y tiempo. El objetivo de este trabajo es la evaluación de un método físico para la extracción rápida del ADN, a partir de colonias mediante el calentamiento y la congelación, para lo cual se evaluaron dos temperaturas 100oC y 95oC. Los resultados mostraron que la extracción de ADN a partir de colonias a 100oC es válida para la genotipificación rápida de S.suis fácilmente en corto tiempo, mientras la temperatura de 95oC no fue suficiente para la liberación del ADN. El límite de detección del ensayo a partir de ADN genómico extraído por purificación química fue 0.5 ng; teniendo en cuenta el tamaño del genoma de S. suis. Es posible considerar que en una simple colonia de S.suis existe la suficiente cantidad de células para garantizar la sensibilidad del ensayo de PCR.
Palabras clave: Streptococcus suis, PCR directo de colonia.
Streptococcus suis is an important pathogen for pigs worldwide. This microorganism is associated with meningitis, arthritis, endocarditis, septicemia, pneumonia and sudden death in pigs during post-weaning and growing (1,2,3). S. suis is also associated with human infections, and is considered an occupational hazard for abattoir workers, meat workers and veterinarians (4,5,6,7 ). S.suis is a diverse species, approximately 33 serotypes of this entity have been described with differences in pathogenicity and geographic distribution, which can be detected by agglutination with the specific antiserum (8,9,10,11,12,13) and also by amplification of fragments of genes related to the capsule polysaccharide biogenesis (14,15). Serotype 2 strains are considered to be highly virulent based on European and Asian epidemiological studies or experimental infections (15,16).
Several molecular tests have been developed to detect S. suis species by means of regions conserved in all the capsular types. Okwumabua et al. (18) developed a PCR assay based on the gdh gene, which encodes the glutamate deshydrogenase, and Marois et al (19) developed a PCR system for S. suis detection by amplifying a fragment of RNAr16s.
Nucleic acid based tests are increasingly used in the bacteriological diagnosis for the speed, sensitivity and specificity, which exceed the benefits of the identification by biochemical tests (18). The isolation and purification of DNA is a key step for most protocols in molecular biological studies including PCR. The various methods proposed to extract and purify DNA from bacterial and yeast can be classified according to the system chosen to break the cells, including beadbeating, enzymatic cell wall lysis or cell permeabilization with chaotrophic agents; generally all the systems either are very time-consuming or they show poor release of DNA (20). The application of a direct PCR from colonies was first performed in rapid characterization studies of Escherichia coli strains transformed with plasmids (21). The DNA amplified directly from the colony has been sequenced with as satisfactory results as those obtained from DNA extracted by the conventional phenol-chloroform procedure (22, 23, 24). The aim of this study was to evaluate a method for the rapid release of the genomic DNA from S.suis colony by using a physical method based on heating and freezing; in this case, two heating temperatures, 100oC and 95oC, were tested for analyzing by polymerase chain reaction.
A total of 10 isolates of S.suis from lungs of pigs with respiratory disorders were cultured on Columbia agar base (Oxoid) supplemented with 5% sheep blood and they were identified with the following criteria: presence of pinpoint colonies with alpha-hemolysis, Gram-positive cocci, negative catalase test and biochemical tests API 20 STREP kit (BiomeÂrieux, Marcy-l'Etoile, France). The conventional phenol-chloroform DNA extraction, followed by ethanol precipitation according to the protocol reported by Douglas et al (25), from overnight broth cultures was used as the control.
For the rapid direct colony PCR, two protocols were followed. The first one consisted of lightly touching a colony of a culture on blood agar with a sterile pipette tip and placing of the collected material into a tube containing 50 µL nuclease-free water, then subjected to boiling at 100oC for five minutes and subsequently frozen at -20oC for 10 minutes, the mixture was centrifuged at 3000 g for 10 minutes. In the second procedure the colony was preheated at 95oC for 10 minutes in the thermal cycler and cooled. In both cases, 5 µL was used for PCR amplification. A colony from both 24 and 48 hour cultures were used. Finally, the mix was added into the two sets of samples separately.
The PCR with extracted genomic DNA as template of strains of S. suis was made as reported by Marois et al. (19). The primers amplify a fragment of 294 bp of 16S rRNA gene. For genoserotyping, specifically the gene fragment related to the biogenesis of the two capsular type (cps2), the conditions described by Smith et al (26) were followed.
Amplification was performed in a final volume of 25 µL containing 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 3 mM MgCl2; 0.1mg/mL BSA; 10 mM of each dNTP; 20 pmol of each primer and 1 µL of Amplicen (2µ/µL), CENSA, Cuba, and 2µl of template DNA, purified by the method of chemical analysis, was added. Different concentrations of the genomic DNA from one isolate were made to establish the detection limit of the assay, and 5 µL was used for PCR amplification for the DNA extracted from the colony.
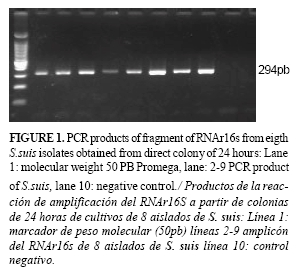
All the isolates of S.suis, identified by morphological and biochemical criteria, amplified a fragment of 294 bp from the genomic DNA extracted by the method of chemical lyses. The amplification limit of the PCR corresponded to 0.5 ng of chromosomal DNA.
In both cases, colony from 24 and 48 hour cultures, the application of direct colony PCR was successful only when the samples were subjected to boiling at 100oC (Figure 1) but not when heating at 95oC, this latter condition was not sufficient for DNA release . Taking into account the detection limit detected from the genomic DNA (0.5 ng), considering the size reported for S. suis genomic DNA (2.14Mpb), assuming that the genome is of GC%=50, then it is possible to estimate a detection limit corresponding to about 2.1x104cells (27). A single 24 hour colony grown on an agar plate contains the number of cells required for PCR amplification of fragment RNAr16s gene and locus fragment linked to biogenesis specific two capsular polysaccharide type.
DNA extraction from Gram-positive bacteria may be more complex than from Gram-negative bacteria and involves multiple steps such as cell wall treatment with enzymes or ionic detergents and cell lysis using mutanolysin and hyaluronidase. These methods are costly, time consuming and often lead to errors when processing a large number of samples (25, 26). For S. suis genotyping, methods based on chemical purification which include the use of proteinase K, detergents such as Triton X-100, Nonidet P-40 and washing with phenol and chloroform are described (28). Trudy et al (29) reported the detection of genes in S. pneumoniae from a colony which was subjected to a chemical lysis solution and heated at 60oC for one hour or at 95oC for 5 minutes.
However, in recent years, several have been the reports on the use of PCR after the rapid extraction of DNA from the colony of Gram-positive bacterial entities. Boiling of the samples has been shown to be a simpler and more economical method for releasing DNA from bacteria (29). The rapid detection of Staphylococcus aureus resistant to methicillin was made from the colony DNA without the use of chemical reagents, but, despite the larger size of the single colony of S. aureus, 4 to 5 colonies were used,(29). Okwumabua et al. (18) used lysis by a boiling method for the PCR assay using gdh gene of S. suis. Briefly, a single colony of a bacterial isolate grown on sheep blood agar plate was suspended in 100 µl of water and heated at 100oC for 20 min, followed by centrifugation for 2 min at 13 000g. However, in our study, the use of a singe colony of 24 hours heated at 100oC for 5 min is enough for the application of the PCR test for genotyping S.suis using different genetic markers. This colony is touched with a simple sterile pipette tip; previously, and using the same pipette tip, this colony is placed on a sheep blood agar plate to be sub-cultured for other assays like antibiotic susceptibility and for ensuring the identity of this colony and its conservation.
Jose and Brahmadathan (28) developed a methodology for the typing of the group A of Streptococcus spp by PCR from colony where preheating at 95oC for 2 minutes and then cooling was sufficient for DNA release. It should be noted that several colonies and not a single one were used in their work. However, our results showed that a colony of S. suis from a culture of 24 hours was enough for typing S.suis.
Although S.suis is a bacterium phenotypically well characterized, its identification in the laboratory may be complicated by the morphological and biochemical similarities with other members of this genus that may be present in the respiratory tract of pigs. Baele et al. (31) studied Gram-positive tonsillar and nasal microbiota in pigs of 2 and 6 weeks of age and identified the following species of Streptococcus spp: S.suis, S. dysgalactiae, S. gallolyticus, S. bovis, S. agalactiae, S. cricetus, S. hyointestinalis, S. hyovaginalis, S. sanguinis, S. porcinus, S. pluranimalium. S.suis was present in all the animals and at concentrations 102 to 107 ufc; however, none of the isolates corresponded to serotype 2. These data demonstrated the need for a rapid protocol for DNA genoserotyping S. suis from mixed primary cultures where other species may be present. The figure 2 shows the amplification products of a fragment of 656 bp of locus cps2j in isolates of S.suis directly from colony.
It was demonstrated that S.suis cells from cultures could be used directly for PCR amplification of target DNA by heating at 100°C for 5 minutes and freezing for 10 minutes at -20°C for cell wall disruption and membrane denaturation; the DNA released was enough for amplification. Thus, these methods can not only replace more cumbersome and time-consuming cell lysate methods, but they also avoids the successive passes needed to obtain pure cultures for the application of biochemical tests and can be used for typing large number of strains in much less time.
REFERENCES
1. Chanter N, Jones PW, Alexander TJ. Meningitis in pigs caused by Streptococcus suis-a speculative review. Vet Microbiol. 1999;36:39-55.
2. Fittipaldi N, Troy EF, Janet FT, Thomas LW. Serotype distribution and production of muramidase-released protein,extracellular factor and suilysin by field strains of Streptococcus suis isolated in the United States. Vet Microbiol. 2009;139:310-317.
3. Gottschalk M, Segura M, Xu J. Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim Health Res Rev. 2007;8:29-45.
4. Thaddeus JW, et al. Slaughterhouse Pigs Are a Major Reservoir of Streptococcus suis Serotype 2 Capable of Causing Human Infection in Southern Vietnam PLoS One. 2011;6(3).
5. Youjun F, Huimin Z, Ying M. George FG. Uncovering newly emerging variants of Streptococcus suis, an important zoonotic agent. Trends Microbiol. 2010;18:3.
6. Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang Hl. Human Streptococcus suis outbreak, Sichuan,China. Emerg Infect Dis. 2006;12:914-920.
7. Kaicheng W, Weixing F, Wisselink H, Chengping L. The cps locus of Streptococcus suis serotype 16: Development of a serotype-specific PCR assay Vet Microbiol. 2011;153: 403-406.
8. Baums CG. Valentin-Weigand P. Surface-associated and secreted factors of Streptococcus suis in epidemiology, pathogenesis and vaccine development. Anim Health Res Rev. 2009;10(1):65-83.
9. Gottschalk M, Higgins R, Boudreau M. Use of polyvalent coagglutination reagents for serotyping of Streptococcus suis. J Clin Microbiol.1993;31(8):2192-2194.
10.Hill J E, Gottschalk M, Brousseau R, Harel J, Hemmingsen SM, Goh SH. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet Microbiol. 2005;107:63-69.
11.Anding Z, Ming Y, Pan H, Jiayan W, Bo C, Yafeng H, et al. Comparative genomic analysis of Streptococcus suis reveals significant genomic diversity among different serotypes. BMC Genomics. 2011;12:523.
12.Andreas B, Katharina B, Christina N, Charlotte S, Karl-Heinz W, Wolfgang B, et al. Comparative evaluation of virulence and pathology of Streptococcus suis serotypes 2 and 9 in experimentally infected growers. Vet Microbiol. 2008;128(3-4):423-430.
13.Lecours Marie-Pier, et al. Sialylation of Streptococcus suis serotype 2 is essential for capsule expression but is not responsible for the main capsular epitope. Microb Infect. 2012;14:941-950.
14.Smith HE, Damamn M, Van der Velde J, Veenbergen V, Wagenaar F, Stockhofe-ZN, et al. Identification and characterization of the complete cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun. 1999;67:1750-1756.
15.Silva LMG, Baums CG, Rehm T, Wisselink HJ, Goethe R, Valentin-Weigand P. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet Microbiol. 2006;155:117-127.
16.Inmaculada L, Verena B, Vera Ana I, Perea JA, Márquez JM, Fernández J, et al . Genetic analysis of Streptococcus suis isolates recovered from diseased and healthy carrier pigs at different stages of production on a pig farm. The Vet Journal. 2010;186:396-398.
17.Zigong W. Characterization of Streptococcus suis isolates from the diseased pigs in China between 2003 and 2007.Vet Microbiol. 2009;137:196-201.
18.Okwumabua O, O'Connor M, Shull E. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol Lett. 2003;218:79-84.
19.Marois C, Le Devendec L, Gottschalk M, Kobisch M. Detection and molecular typing of Streptococcus suis in tonsils from live pigs in France. Can J Vet Res. 2007;71(1):14-22.
20.Mirhendi H, Diba K, Rezaei A, Jalalizand N, Hosseinpur L, Khodadadi H. Colony-PCR Is a Rapid and Sensitive Method for DNA Amplification in Yeasts. Iranian J Publ Health. 2007;36(1):40-44.
21.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.2001.
22.Brian R, Berridge J D, Fuller JA, Donald EL, Herve B, Frelier PF. Development of specific oligonucleotide PCR primers for the Streptococcus iniae 16S-23S ribosomal DNA intergenic spacer. J Clin Microbiol. 1998;36(9):2778-2781.
23.Hynes WL, Ferretti JJ, Gilmore MS, Segarra RA. PCR amplification of Streptoccal DNA using crude cell lysates. FEMS Microbiol Lett. 1992;94:139-42.
24.Hofmann MA, Brian DA. Sequencing PCR DNA amplified directly from a bacterial colony. Biotechniques. 1991;11:30-31.
25.Douglas A, Raúl A, Stephen P. Elucidation of the DNA sequence of Streptococcus uberis adhesion molecule gene (sua) and detection of sua in strains of Streptococcus uberis isolated from geographically diverse locations. Vet Microbiol. 2008; 128(3):304-312.
26.Smith, HE., Veenbergen V, van der Velde J, Damman M, Wisselink HJ, Smits MA. The cps genes of Streptococcus suis serotypes 1, 2, and 9: development of rapid serotype-specific PCR assays. J Clin Microbiol. 1999;37:3146-3152.
27.Takasugu G, Hideake N, Ayko M, Yoshiaki K, Oki O, Kanako A. Rapid identification of Streptococus intermedius by PCR with the ily gene as a specie marker gene. J Med Microbiol. 2002;51;178-186.
28.Jose JJM, Brahmadathan K. Evaluation of simplified DNA extraction methods for EMM typing of group a streptococci Indian. J Med Microbiol. 2006;224(2):127-130.
29.Trudy O, Messmer C, Whitney G, Barry S. Fields Use of polymerase chain reaction to identify pneumococcal infection associated with hemorrhage and shock in two previously healthy young children. Clin Chemistry. 1997;43:930-935.
30.Sandhu GS, Precup JW, Kline BC. Rapid one step characterization of recombinant vectors by direct analysis of transformed Escherichia coli colonies. Biotechniques.1989;7:689-690.
31.Baele M, Chiers K, Devriese LA, Smith HE, Wisselink HJ, Vaneechoutte MF, et al. The Gram-positive tonsillar and nasal flora of piglets before and after weaning. J Applied Microbiol. 2001;91:997-1003.
Recibido: 6-12-2012.
Aceptado: 25-1-2013.