My SciELO
Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista de Salud Animal
Print version ISSN 0253-570XOn-line version ISSN 2224-4700
Rev Salud Anim. vol.38 no.2 La Habana May-Aug. 2016
ORIGINAL ARTICLE
Genotyping of the kappa-casein and leptin genes in Cuban water buffalo by PCR-RFLP
Genotipado por PCR-RFLP de los genes de la kappa-caseína y la leptina en búfalos de agua de Cuba
Nadia Martínez MarreroI, Luciana Amaral de MascenaII, Jamille Lopes de MacedoII, Alina Mitat ValdésIII, Manoel Adrião Gomes FilhoII, Odalys Uffo ReinosaI*
ICentro Nacional de Sanidad Agropecuaria (CENSA). Laboratorio de Genética Molecular. Apartado 10, San José de las Lajas, Mayabeque, Cuba.
IIUniversidade Federal Rural de Pernambuco (UFRPE). Departamento de Morfologia e Fisiologia Animal. Rua DomManoel de Medeiros, s/n, Dois Irmãos - CEP: 52171-900. Recife-PE, Brazil.
IIIUniversidad Agraria de La Habana (UNAH). Facultad de Medicina Veterinaria. Carretera de Tapaste y Autopista Nacional. San José de las Lajas. Mayabeque, Cuba.
ABSTRACT
Genetic analysis of loci affecting production and reproduction traits in livestock animals make possible the evaluation of animal breeding values using molecular markers. The aim of this study was to detect polymorphisms in the leptin and k-casein genes of a Cuban water buffalo population to provide markers useful for associating studies. Genomic DNA from 68 unrelated Cuban buffaloes, uncontrolled crossbreds of river (Buffalypso) and swamp (Carabao) animals, was analyzed using PCR-RFLP. Fragments of leptin and ê-casein genes were amplified and digested with Bsa AI and Hind III/Taq I, respectively. All the amplified samples were monomorphic BB after digestion with Hind III/Taq I. Genotypic frequencies found in the leptin gene with Bsa AI were 0.47 (AA), 0.42 (AG) and 0.11 (GG). The population was in HWE (p=1.0000), possibly because there was not selection for this locus on it. The FIS estimate (0.0212) showed no inbreeding in the population regarding this locus. He (0.436) and Ho (0.426) values were below 0.5 (50%), indicating low genetic variation in this locus in the population. PCR-RFLP detected the two genetic variants described for the leptin gene A1620G. However, a higher number of animals and SNPs in the leptin and/or other genes should be analyzed for a more accurate estimate of the genetic diversity in this population. DNA sequencing should be assayed by nucleotide sequence analysis to detect the new A and B variants of k-casein reported in buffalo.
Key words: leptin, Casein, buffaloes, PCR-RFLP, genetic variants.
RESUMEN
El análisis genético de loci, que controla caracteres productivos y reproductivos, hace posible la evaluación del valor genético en el ganado con el uso de marcadores moleculares. El objetivo de este estudio fue detectar polimorfismos en los genes de la leptina y la k-caseína en búfalos cubanos, para proveer marcadores útiles en estudios de asociación. Se amplificaron por PCR fragmentos de los genes de la leptina y la ê-caseína que se digirieron con Bsa AI y Hind III/Taq I, respectivamente. Todas las muestras amplificadas resultaron monomórficas BB tras la digestión con Hind III/Taq I. Las frecuencias genotípicas en el gen de la leptina fueron, 0.47 (AA), 0.42 (AG) y 0.11 (GG). La población se encontró en equilibrio de Hardy-Weinberg (p=1.0000), posiblemente, porque no está ocurriendo selección para dicho locus. El FIS estimado (0.0212) mostró que no existe endogamia en la población cuando se considera este locus. Los valores de He (0.436) y Ho (0.426) estaban por debajo de 0,5 (50%), lo que indica que existe una baja variabilidad genética para este locus en la población. La PCR-RFLP permitió detectar las variantes genéticas descritas para A1620G en el gen de la leptina. Sin embargo, se deben analizar un mayor número de animales y SNPs, en este u otros genes, para estimar con mayor certeza la diversidad genética en esta población. Es necesario el uso de la secuenciación para detectar las nuevas variantes A y B de la k-caseína que se han descrito en búfalos a través de esta técnica.
Palabras clave: Leptina, Caseína, búfalos, PCR-RFLP, variantes genéticas.
INTRODUCTION
Identification of loci affecting production and reproduction traits in livestock animals would make possible the evaluation of animal breeding values using molecular markers. In genetic animal studies, much attention has been focused on the identification of single nucleotide polymorphism (SNP). These markers are extremely common throughout the genome and their identification is the first step in association studies (1).
Many studies in bovine have reported association between SNPs and traits of economic interest (2, 3, 4). Genetic polymorphism in milk proteins has raised great interest in the animal breeding and dairy industry, due to the relationship between milk proteins and milk production traits, composition, and quality (5, 6, 7). Kappa-casein (k-casein, CSN3, K-Ca, kCn, CASK) is one of the most important and highly studied milk protein genes. In cattle 13 protein variants and one synonymous variant have been reported; however, the most frequent ones are A and B alleles (8). The variants of k-casein affect casein content, protein content and cheese yield, as well as curd firmness.
The leptin gene is another important candidate gene that can be used to improve animal reproduction and production. Leptin and its receptor have been mapped in several species and a number of SNPs have been identified for further use in marker assisted selection programs (9). Polymorphism in the bovine leptin gene locus associated with genetic variation in energy balance, milk production, live weight, and fertility trait have been reported by many researchers (10, 11, 12).
Before the use of these genes for enhancing productivity and reproduction in farm animals, studies with different populations are required for a proper characterization of the robustness of their association with economically important traits across dairy/beef livestock (13). Effective genotyping of the k-casein and leptin genes of buffaloes requires fast, efficient, and low cost methods, independent of age and sex. DNA based genotyping through polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) has made this evaluation possible as well as simple for a large number of animals (14). Therefore, the aim of the present study was to detect some SNPs, previously reported in cattle, in the leptin and k-casein genes of a Cuban water buffalo population by PCR-RFLP to provide markers useful for association studies in this population.
MATERIAL AND METHODS
Genomic DNA isolation
Blood samples were collected from 68 unrelated Cuban buffaloes, uncontrolled crossbreds of river (Buffalypso) and swamp (Carabao) buffaloes belonging to a dairy herd at the Empresa Pecuaria Genética «El Cangre», in Mayabeque Province.
Genomic DNA was extracted using Promega Wizard® Genomic DNA purification kit (Promega, Madison, WI, USA) in accordance with the manufacturer's suggested protocol.
The quality and quantity of DNA (ng/µL) for each sample were analyzed in a spectrophotometer (Nanodrop ND1000, Thermo Scientific).
PCR amplification
A fragment of 522 bp located between intron 2 and exon 3 of the leptin gene was amplified by PCR using the primers 5´-GTC TGG AGG CAA AGG GCA GAG T-3´ and 5´-CCA CCA CCT CTG TGG AGT AG-3´ (15). The reaction mixture contained ~100 ng of genomic DNA, 0.2 mM of dNTPs, 2mM of MgCl2, 0.4 µM of each primer, 1X of Taq Platinum DNA Polymerase Buffer, 0.75 U of Platinum®Taq DNA polymerase (Invitrogen™/Life Technologies, Carlsbad, CA, USA), and nuclease free water (Promega, Madison, WI, USA) to make a final volume of 25 µl. Amplification conditions were as follow: 95°C for 5 min, 30 cycles at 94°C for 15sec, 65°C for 30 s and 72°C for 1 min, followed by a final extension step of 72°C for 5 min.
A 379 bp fragment from exon IV of k-casein gene was amplified using the primers K-F: 5´-CACGTCACCCACACCCACATTTATC-3´ and K-R: 5´-TAATTAGCCCATTTCGCCTTCTCTGT-3´ (17). The PCR mixture was composed of ~100 ng of genomic DNA, 1XPCR master mix (Promega, Madison, WI, USA), 0,4 µM from each primer, 2 mM of MgCl2, and nuclease free water (Promega, Madison, WI, USA) to make a final volume of 25 µl. The PCR reaction included pre-denaturation for 4 min at 95°C followed by 30 cycles at 94°C for 1 min, 56°C for 2min, 72°C for 1 min and a final extension of 7 min at 72°C.
The amplified products were stained with Blue Green Dye (LGC Biotecnologia, Cotia, SP, Brazil) and electrophoresed in agarose gel (2 %) for 45 min at 100 V in 0.5 X TBE buffer. The visualization was done under ultraviolet light and the gel was documented by Gel-Doc (Bio-Rad) equipment. The presence of the expected bands and the quantity of PCR products were assessed by comparing with a 100 bp DNA ladder (Promega, Madison, WI, USA).
Genotyping using RFLP
The PCR products of the leptin gene amplification were digested with Bsa AI restriction enzyme to discriminate A or G allelic variants (15, 16). The restriction digestion was performed in a total volume of 15 µl containing 1X of enzyme buffers (ThermoScientific, Waltham, MA, USA) 0.1-0.5 µg of amplified DNA, 10 U Bsa AI (ThermoScientific, Waltham, MA, USA), and nuclease free water (Promega, Madison, WI, USA). The reaction mixture was incubated at 30°C for 3 h.
In the case of the k-casein gene, the 379 bp PCR products were digested with Hind III or Taq I restriction enzymes to discriminate A or B allelic variants (14). The restriction digestions were performed in a total volume of 25 µl containing 1X of enzyme buffers (Promega, Madison, WI, USA), 0.1-0.5 µg of amplified DNA, 10 U Hind III or Taq I (Promega, Madison, WI, USA), and nuclease free water (Promega, Madison, WI, USA). The reaction mixture was incubated at 37°C (Hind III) or 65°C (Taq I) for 3 h.
The restriction products were stained with Blue Green Dye (LGC Biotecnologia, Cotia, SP, Brazil) and electrophoresed in agarose gel (3 %) for 45 min at 100 V in 0.5 X TBE buffer. The visualization was done under ultraviolet light and the gel was documented by Gel-Doc (Bio-Rad) equipment. The size of the bands was estimated with a 50 bp (leptin gene) or 100 bp DNA ladder (Promega, Madison, WI, USA).
Statistical Analysis
Direct counting was used to estimate genotype frequencies of gene genetic variants.
Deviations from Hardy-Weinberg Equilibrium (HWE) and Wright Fixation Index (FIS, within population inbreeding estimate) were assed with GENEPOP v4.2 software (17) using exact tests that employ the Markov Chain method to estimate p-values (1000 dememorization steps, 100 batches and 1000 iterations). Allele frequencies and observed (Ho) and unbiased expected (He) heterozygosities were calculated using the Excell complement GenAlEx 6.5 (18).
RESULTS
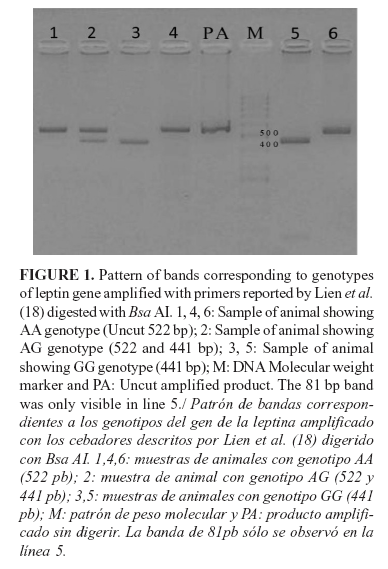
All 68 samples were amplified using specific primers for the leptin gene. After digestion of PCR products with Bsa AI, an intact 522 bp fragment as AA genotype, 441 and 81 bp fragments as GG genotype, and 522, 441 and 81 bp fragments as GA genotype were observed (Figure 1). Genotypic frequencies were 0.47 for genotype AA, 0.42 for AG, and 0.11 for GG. The allelic frequencies found were 0.684 for the allele A and 0.316 for the allele G. The population was in HWE (p-value =1.0000). FIS estimates were 0.0212. He value was 0.436, and Ho was equal to 0.426.
Sixty-five samples were amplified with primers K-F and K-R for exon IV of k-casein. After digestion of PCR products with Hind III, all animals were monomorphic, showing two fragments of 225 and 154bp corresponding to BB genotype (Figure 2). The 379bp amplified a fragment remained undigested by Taq I restriction enzyme in all animals, also showing a monomorphic BB pattern.
DISCUSSION
It has been demonstrated by interspecies comparisons that k-casein possesses the highest degree of conservation among the casein genes (19). The leptin gene structure, intron/exon boundaries and amino acid sequence are highly conserved in mammalian species. Comparative study of intron 2 of the leptin gene between bovines, buffaloes and goats, revealed the sequence similarity of 95-98% and indicated that bovine were closer to buffaloes (10). The former explains that buffalo leptin and k-casein genes were successfully amplified by PCR using primers designed for bovines. Failure of amplification in three samples in the k-casein gene could be due to presence of PCR inhibitors in the DNA sample or because of the existence of mismatches in the union site of primers in the gene sequence.
Lien et al. (15) first described a guanine (G) to adenine (A) substitution in position 1620 in intron 2 of the leptin gene of Norwegian cattle identified by Bsa AI digestion. Chaudhary et al. (20) reported digestion of 522 bp PCR products with the Bsa AI restriction enzyme and revealed three genotypes in all the breeds of Bos indicus, Bos taurus, and Jersey cattle. Souza et al. (5) determined leptin A1620G to be associated with weaning weight (W) in three Nelore (Bos indicus) lines selected for growth. Significant effects (p=0.03) of this polymorphism were observed for W210, with the demonstration of greater mean values for AA animals compared with AG and GG animals. There have been few studies on this SNP in water buffaloes.
The results of the present study in a Cuban buffalo population showed the presence of the three genotypes (AA, AG, and GG), with AA as the most frequent genotype. The genotype GG was the least frequent. In accordance with that fact, allele A showed the highest allele frequency. The population in HWE indicated that, possibly, there was not selection for this locus in the population. FIS estimates showed that no inbreeding occurred in the population regarding this locus.
Heterozygosity value is the most accurate way to measure the genetic diversity of population (21) and to get an overview of the genetic variability (22). He and Ho values obtained in this study for the leptin gene were below 0.5 (50%). Javanmard et al. (23) suggest that heterozygosity values below 0.5 (50%) indicate low variation of a gene in the population. The number of samples, the number of alleles, and the allele frequencies influence heterozygosity values. A higher number of animals and SNPs in the leptin and/or other genes should be analyzed for more accurate estimate FIS and genetic variation in this population.
Comparing with other researches made on water buffalo, Azari et al. (24) analyzed 53 Mazandarani native river buffaloes from Iran by PCR-RFLP in leptin gene using Bsa AI. They also observed three genotypes. The highest genotype frequency detected by the authors corresponded to AG (AB) (0.509) genotype, different from the results of the present study. They reported frequency of GG (BB) genotype as the lowest, in accordance with this work. The allelic frequencies that Azari et al. (24) reported for A and G (B) alleles were 0.61 and 0.35, respectively. The first one was higher, and the second lower than the allelic frequencies detected in the present study. The Mazandarani buffalo population was in HWE for the locus analyzed as was observed in the Cuban population. The Ho (0.509) was slightly superior to that observed in this study but it was still near to 0.5.
In another research, Zetouni et al. (16) studied a Brazilian buffalo population also by PCR-RFLP in the leptin gene using Bsa AI. The authors identified the three genotypes and reported the highest frequency for AG (0.54) genotype and the lowest for GG (0.16). The allele frequency they found for the A allele (0.57) was lower than that observed in this work and the allele frequency for the G allele (0.43) was higher. The author informed that the population was in HWE for this locus as the population study in this research. The Ho (0.54) was higher than that obtained in the present work.
Kale et al. (25) reported that digestion of the 522 bp fragment of Murrah buffalo leptin gene with Bsa AI yielded an uncut fragment, which indicated the frequency of AA genotype as 1. The polymorphic restriction site was absent in the Murrah buffaloes studied exhibiting monomorphic pattern in the analyzed population.
The analyzed Cuban buffalos resulted from uncontrolled crossbred of Carabao and Bufallypso, which is a composite breed formed by Murrah, Surti, Jaffarabadi, Nelli and Bhadawari (26). Considering the results reported by Kale et al. (25), the contribution made by the Murrah Indian riverine breed to the formation of Buffalypso could explain why AA was the most frequent genotype in the studied Cuban population.
In exon IV of cattle k-casein gene, alleles A and B were detected by differences at codons 136 and 148 (27). Allele A has 136Thr (ACC)/148Asp (GAT), where as allele B has 136Ile (ATC)/148Ala (GCT). All previous reports on genotyping of water buffalo k-casein gene (exon IV) by PCR-RFLP analysis used restriction enzymes Hind III, Hinf I, and Taq I, which successfully identify cattle A and B genotypes. The results obtained in the analysis of Cuban buffalo k-casein gene were in accordance with Mitra et al. (14), who reported monomorphism (BB) for this gene, showing similar band pattern of 225 bp and 154 bp by using restriction endonuclease Hind III in Murrah and Nili-Ravi buffalo breeds. In other researches using PCR-RFLP, Pipalia et al. (27), Riaz et al. (28), Abbasi et al. (29), Ren et al. (30), and Jaayid et al. (31) also found monomorphism (BB) for this gene in diverse buffalo breeds from different countries.
However, Sing et al. (32) found the two alleles A and B for k-casein locus in Murrah and Bhadawari breeds by PCR-RFLP, but they reported monomorphism (BB) in Surti and Mehsana breeds of buffalo. Similarly, Patel et al. (33) found alleles A and B in Murrah, Surti, and Pandharpuri buffaloes. They observed BB and AB genotypes in these three breeds and monomorphism (BB) in Jaffarabadi breed. The authors found BB genotype frequency (0.968) very much higher than the AB genotype (0.032). In another work, Gouda et al. (34) reported detection of BB and AB genotypes in Egyptian buffalo, also with higher frequency of BB genotype (0.75). Lin et al. (35) also studied a sample from different breeds and crossbreds of buffalo in China by RFLP. They found the three k-casein genotypes AA, AB, and BB, though the latter genotype occupied the largest proportion. The authors detected that most of the AB genotype and all of the genotype AA appeared in crossbred buffalo (Murrah + Nili-Ravi and river type + Chinese local swamp), while just several AB and none of AA genotype were found in pure river type buffalo (Murrah, Nili-Ravi).
In the present study, a population of uncontrolled crossbred animals (composite river breed + swamp) was analyzed. Taking into account the presence of alleles A and B for k-casein locus in Murrah reported by Sing et al. (32) and Patel et al. (33), and the presence of AA and AB genotypes in crossbred buffalo (Murrah + Nili-Ravi and river type + Chinese local swamp) and AB genotype in pure Murrah, referred to by Lin et al. (35), it was expected that Cuban buffaloes showed a polymorphic pattern in k-casein exon IV by PCR-RFLP. However, the results are not in agreement with those of all these authors and agree with those obtained by Otaviano et al. (36), who also found BB monomorphism in k-casein exon IV when they studied Brazilian Murrah breed and its crossbreds by PCR-RFLP.
Buffalo k-casein polymorphism has also been investigated using nucleotide sequence analysis, and two nucleotide variants at codons 135Thr (ACC)/Ile (ATC), and 136Thr (ACC/ACT) (silent mutation) have been reported (14, 37-39). Allele A has 135ThrACC/136ThrACC, whereas allele B has 135IleATC/136ThrACT. In comparing partial nucleotide sequences (from codon 130 to 149) of alleles A and B in cattle and buffalo, Nahas et al. (39) found that Hind III restriction site ``A^AGCTT'' was present in cattle allele B (148Ala) and in both buffalo alleles A and B, whereas Hinf I restriction site ``G^ANT'', contrary to Hind III, was present in cattle allele A (148Asp) and was missing in cattle allele B as well as in both buffalo alleles A and B. Since buffalo samples (both alleles A and B) followed cattle BB pattern, they were mistakenly assumed as BB monomorphic where in fact they would have been AA, BB or AB. Taq I restriction site T^CGA is present at codon 136 in cattle allele B (136Ile) but was absent in buffalo alleles A and B. These findings are in contrast with those results referred to by Sing et al. (32), Patel et al. (33), Gouda et al. (34) and Lin et al. (35).
PCR-RFLP technology has been extensively used in buffalo DNA analysis (14, 16, 20, 25, 31, 40-43). In this study, PCR-RFLP with Bsa AI was able to detect the two genetic variants described for the leptin gene A1620G. However, the monomorphism BB finding in k-casein gen using Hind III and the absent of a Taq I restriction site, indicates the necessity of DNA sequencing, as a method with higher specificity and accuracy (44), to detect the new A and B variants of k-casein reported in buffalo by nucleotide sequence analysis (45, 47, 48). As it was referred by Caroli et al. (44), when funding is limited, researchers may use RFLP analysis for screening of the total sample set for polymorphism, after which, samples exhibiting different RFLP patterns may be subjected to DNA sequencing analysis.
REFERENCES
1. Anand VK, Sharma V, Kumar A, Verma VK, Rana VP, Khirbat R, et al. PCR-RFLP based leptin gene polymorphism and its association with mastitis in Murrah buffalo. J Cell Tissue Res. 2015;15(2):5125-5132.
2. Alashawkany AR, Shahroudi FE, Nassiry MR, Moussavi AH, Heydarpour M, Sadeghi B. Association of SNP in the Exon II of Leptin gene with Milk and Reproduction Traits in Holstein Iranian Cows. Biotechnology. 2008;7:347-350.
3. Deb R, Singh U, Kumar S, Singh R, Sengar G, Sharma A. Genetic polymorphism and association of kappa-casein gene with milk production traits among Frieswal (HF × Sahiwal) cross breed of Indian origin. Iran J Vet Res. 2014;15(4):406-408.
4. Schenkel FS, Miller SP, Jiang Z, Mandell IB, Ye X, et al. Association of a single nucleotide polymorphism in the calpastatin gene with carcass and meat quality traits of beef cattle. J Anim Sci. 2006;84:291-299.
5. Souza FRP, Mercadante MEZ, Fonseca LFS, Ferreira LMS, Regatieri IC, Ayres DR, et al. Assessment of DGAT1 and LEP gene polymorphisms in three Nelore (Bos indicus) lines selected for growth and their relationship with growth and carcass traits. J Anim Sci. 2010;88:435-441.
6. Caroli A, Chessa S, Bolla P, Budelli E, Gandini GC. Genetic structure of milk protein polymorphisms and effects on milk production traits in a local dairy cattle. J Anim Breed Genet.2004;121:19-127.
7. De Marchi M, Bittante G, Dal Zotto R, Dalvit C, Cassandro M. Effect of Holstein Friesian and Brown Swiss breeds on quality of milk and cheese. J Dairy Sci. 2008;91:4092-4102.
8. Gallinat JL, Qanbari S, Drögemüller C, Pimentel ECG, Thaller G, Tetens J. DNA-based identification of novel bovine casein gene variants. J Dairy Sci. 2013;96:699-709.
9. Gallegos MP, Toca JA, Rodriguez P, Pinzón CE, Reveles FA, Quintero JS. Role of leptin gene in cattle production: review. J Anim Vet Adv. 2015;14(4):81-90.
10.Argawal P, Rout PK, Singh SK. Leptin: A biomolecule for enhancing livestock productivity. Indian J Biotechnol. 2009;8:169-176.
11.Nkrumah JD, Li C, Yu J, Hansen C, Keisler DH, Moore SS. Polymorphisms in the bovine leptin promoter associated with serum leptin concentration, growth, feed intake, feeding behavior, and measures of carcass merit. J Anim Sci. 2005;83:20-28.
12.Haegeman A, Van Zeveren A, Peelman LJ. New mutation in exon 2 of the bovine leptin gene. Anim Genet. 2000;31:79.
13.Datta S, Adikari AMJB, Chauhan A, Verma A, Gupta ID, Chauhan I, et al. Nucleotide sequence variation in leptin gene of Murrah buffalo (Bubalus bubalis). Exper Anim Med Res. 2012;2:130-136.
14.Mitra AP, Schlee I, Krause J, Blusch T, Werner CR, et al. Kappa casein polymorphism in the Indian dairy cattle and buffalo: A new genetic variant in buffalo. Anim Biotechnol. 1998;9:81-87.
15.Lien S, Sundvold H, Klungland H, Vage DI. Two novel polymorphisms in the bovine obesity gene (OBS). Anim Genet. 1997;28:45.
16.Zetouni L, de Camargo GMF, Fonseca PDS, Monsalves FMG, Hurtado NA, Aspilcueta-Borquis RR, et al. Effects of a single nucleotide polymorphism in the Leptin gene on the productive traits of dairy buffaloes (Bubalus bubalis). Mol Biol Rep. 2013;40:5159-5163.
17.Rousset F. Genepop'007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour. 2008;8:103-106.
18.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research - an update. Bioinformatics. 2012;28:2537-2539.
19.Ahmed TJ, Yakoub MY, Falih BZ, Mohammed JO. Genetic Polymorphism of Kappa Casein (k-CN) in Iraqi Buffalo Using Polymerase Chain Reaction-Restriction Fragment Length Polymorphism. J Agr Sci Tech. 2013;835-841.
20.Choudhary V, Kumar P, Bhattacharya TK, Bhushan B, Sharma A. DNA polymorphism of Leptin gene in Bos indicus and Bos taurus cattle. Genet Mol Biol. 2005;28:740-742.
21.Nei M, Kumar S. Molecular Evolution and Phylogenetics. 2000. Oxford University Press, New York.
22.Marson EP, Ferraz JBS, Meirelles FV, Balieiro JCDC, Eler JP, et al. Genetic characterization of European Zebu composite bovine using RFLP markers. Genet Mol Res. 2005;4:496-505.
23.Javanmard A, Asadzadeh N, Banabazi MH, Tavakolian J. The allele and genotype frequencies of bovine pituitary specific transcription factor and Leptin genes in Iranian cattle and buffalo populations using PCR-RFLP. Iranian J Biotech. 2005;3:104-108.
24.Azari MA, Hasani S, Heidari M, Yousefi S. Genetic polymorphism of Leptin gene using PCR-RFLP method in three different populations. Slovak J Anim Sci. 2012;45:39-42.
25.Kale DS, Yadav BR, Mukherjee A, Prasad J. Exploring DNA Polymorphisms of Leptin Gene within Indian Water Buffaloes. J Adv Vet Res. 2013;3:20-26.
26.Robitaille G, Britten M, Morisset J, Petitclerc D. Polymorphism in the bovine kappa-casein (CSN3) gene and the 50-flanking region: sequence analysis of CSN3 A and B alleles. J Anim Genet. 2005;36:184-185.
27.Pipalia D, Ladani D, Brahmkshtri B, Rank D, Joshi C, et al. Kappa-casein genotyping of Indian Buffalo breeds using PCR-RFLP. Buffalo J. 2001;17:195-202.
28.Riaz MN, Malik NA, Nasreen F, Qureshi JA. Molecular marker assisted study of kappa-casein gene in Nili-Ravi (buffalo) breed of Pakistan. Pakistan Vet J. 2008;28:103-106.
29.Abbasi B, Fayazi J, Beigi Nasiri MT, Roshanfekr HA, Mirzadeh Kh, Sadr AS. Analysis of kappa casein gene polymorphism by PCR-RFLP in Buffalo population in Khouzestan Province. Res J Biol Sci. 2009;4:1073-1075.
30.Ren DX, Miao SY, Chen YL, Zou CX, Liang XW, Liu JX. Genotyping of the k-casein and b-lactoglobulin genes in Chinese Holstein, Jersey and water buffalo by PCR-RFLP. J Genet. 2011; 90: e1-e5. (http://www.ias.ac.in/jgenet/OnlineResources/90/e1.pdf).
31.Jaayid TA, Muntaha YY, Bashar FZ, Jaafer M. Genetic Polymorphism of Kappa Casein (b-CN) in Iraqi Buffalo Using Polymerase Chain Reaction-Restriction Fragment Length Polymorphism. J Agric Sci Technol. 2013; 835-841.
32.Singh S, Pushpendra K, Bhattacharya TK. DNA polymorphism of k-CN and b-CN genes and its association with milk production and quality traits in buffalo (B. bubalis). Proceeding of National Symposium on Domestic Animals Diversity: Status, Opportunities and Challenges, 2005. Karnal, India.
33.Patel JB, Chauhan KMS, Soni KJ. Genotyping and allelic frequencies of k-CN and b-LG in Indian river buffalo bulls. Buffalo Bull. 2007;26:63-66.
34.Gouda EM, Galal MKh, Abdelaziz SA. Genetic Variants and Allele Frequencies of Kappa Casein in Egyptian Cattle and Buffalo Using PCR-RFLP. J Agric Sci. 2013;5:197-203.
35.Lin B, Ren D, Yang B, Li L, Yan T, et al. Compare of Phenotypic Variation with the Polymorphism of One Coagulation Related Gene Locus in Buffalo Kappa-Casein. Proceeding of the 10th World Buffalo Congress 2013. Puket, Thailand.
36.Otaviano R, Tonhati H, Desiderio JA, Munoz FC. Kappa-casein gene study with molecular marker in female buffalos. Genet Mol Biol. 2005;28:237-241.
37.Beneduci B, Krastanov I, Maretto F, Oblakov N, Cassandro M. Molecular characterization of ê-casein allelic variants in Bulgarian buffalo breed. Acta Agraria Kaposvariensis. 2010;14:169-172.
38.Bonfatti V, Giantin M, Gervaso M, Coletta A, Dacasto M, Carnier P. Effect of CSN1S1-CSN3 (aS1-k-casein) composite genotype on milk production traits and milk coagulation properties in Mediterranean water buffalo. J Dairy Sci. 2012;95:3435-3443.
39.Nahas SM, Bibars MA, Taha DA. Genetic characterization of Egyptian buffalo CSN3 gene. J Genet Eng Biotechnol. 2013;11:123-127.
40.Jhala NB, Vataliya PH, Rank DN, Ramani UV. Leptin gene exon-3 polymorphism in Gir cattle and Mehsana buffalo. J Anim Sci Reporter. 2011;5:91-94.
41.Somayeh R, Vida K, Sadr A, Moosavi M. Investigation of leptin polymorphism by PCR-RFLP in Iranian buffalo. Intl Res J Appl Basic Sci. 2012;3(8):1658-1661.
42.Rini AO, Sumantri C, Damayanthi E. ê-casein gene polymorphisms in riverine and swamp buffalo in Indonesia. J Indonesian Trop Anim Agric. 2014;39:1-9.
43.Martin ET, Koelle DM, Byrd B, Huang ML, Vieira J, et al. Sequence-based methods for identifying epidemiologically linked herpes simplex virus Type 2 strains. J Clin Microbiol. 2006; 2541-2546.
44.Caroli A, Chiatti F, Chessa S, Rignanese D, Bolla P, Pagnacco G. Focusing on the goat casein complex. J Dairy Sci. 2006;89:3178-3191.
Recibido: 5-1-2016.
Aceptado: 3-6-2016.
*Corresponding author: Odalys Uffo Reinosa. E-mail: uffo@censa.edu.cu