Introduction
Leptospirosis is a zoonosis of worldwide distribution with great importance in public health. The disease is caused by pathogenic bacteria of the genus Leptospira, whose reservoirs are domestic and wild animals.1
Leptospirosis is associated with natural disasters, floods, and hurricanes as well as with a precarious infrastructure and unfavorable socioeconomic conditions, such as areas on the banks of rivers that are occupied by people who are displaced due to social and political reasons and areas with improper waste disposal. These environments favor increased populations of dogs and rodents that are primary reservoirs for leptospira in urban and rural areas.2,3,4
Wild rats (Rattus spp.), the brown rats (Rattus norvegicus), and the black rats (R. rattus) are abundant in peri-domestic environments and constitute asymptomatic reservoirs for different serovars of Leptospira interrogans, which colonize their renal tubules and can cause the disease in other animals and humans.2,5,6
Different values of seroprevalence of the disease have been reported in Colombia; for example, a study carried out in Urabá Antioqueño included 479 patients with acute febrile illness, 58% of which (278/479) had a confirmed diagnosis of leptospirosis.7 Serum of 62 workers from a pig farm in the middle Sinú department, Córdoba, was analyzed, and 75.80% (n=47) of them had antibodies against L. interrogans (sensu lato).8 In Bogotá, there was a seroprevalence of 12.6% (165/1307),9 where as Tolima reported the lowest seroprevalence in the country with 6% (51/850).10
The municipality of Villavicencio is the capital of the department of Meta and is located at 04° 09 N, 73° 38 W and has a population of approximately 451,212 inhabitants.11 It has an altitude of 467 meters above sea level and an average temperature of 30°C. The municipality has large physiographic areas, a feature that favors the development of agricultural, livestock, agro-industrial, agro-tourism, and ecotourism activities that currently account for the main economic activities in the region. The vegetation of the plain mainly consists of pastures and grasslands with abundant shrubs and low trees. Sampling included peri-urban and rural areas of townships belonging to the municipality of Villavicencio, which shared similar ecological conditions, being peri-domiciliary areas close to water bodies. Some of them presented unfavorable sanitary conditions and others were fields of typical crops of agricultural areas.
The department of Meta has a population close to one million inhabitants and meets all the climatic, environmental, sanitary, and social conditions for the development of the disease, in addition to being endemic for other tropical diseases that are included in the category of acute febrile illness. In 2018, Sánchez et al., found that of 100 patients with acute febrile illness, 29% were diagnosed with leptospirosis using the microagglutination test, with Canicola and Ballum being the most prevalent serovars.12) In the department of Meta, leptospirosis is underreported as regional laboratories lack the techniques required to diagnose the disease and have focused on diagnosing dengue, which negatively impacts the local and national epidemiological surveillance system.
Previous studies in Villavicencio reported that pigs taken to the slaughterhouse were reservoirs for various serogroups of leptospira. However, no studies have been carried out on wild or synanthropic rodents from rural and peri-urban areas of the municipality; therefore, this study aimed to detect the presence of Leptopsira spp. through a molecular analysis in rodents (Rodentia) from peri-urban and rural areas of the municipality of Villavicencio in Colombia.
Methods
Capture of rodents
A total of 60 Sherman-type traps were set out in 7 townships of the municipality of Villavicencio and placed in strategic sites where there was waste accumulation, food storage, or agriculture. The bait used was a mixture of flaked oats with banana and peanut butter. The traps were left overnight and checked early the next morning, in the same place, for 7 days. Trapping was carried out on 10 occasions starting in October 2018 and ending in October 2019.
Biological sampling
For morphometric identification, the rodents were anesthetized using 0.1 or 0.2 ml of 10% ketamine hydrochloride depending on their weight. Then a cardiac puncture was performed to extract blood, and they were later euthanized under anesthesia. The organs extracted were labeled and placed into cryovial tubes and subsequently preserved in liquid nitrogen.13 A mammalogist with experience in the area conducted the identification of the species.
DNA extraction
Total DNA was extracted directly from kidney tissue using the commercial GeneJET Purification Kit (K0722) from ThermoFisher Scientific following the recommendations of the supplier. The purified DNA was stored at −20°C until use. DNA quantification was performed in a NanoDrop™ 2000 spectrophotometer with optical densities of 230, 260, and 280 nm to calculate the 260/280 ratios to obtain the concentration and purity of the DNA of the samples.
Polymerase chain reaction (PCR) and sequencing
The DNA extracted from the kidney was subsequently amplified through conventional PCR. The following primers were used: pfLp32-1 5´-TAGAATCAAGATCCCAAATCCTCC-3´ and pfLp32-2 5´-CCAACAGATGCAACGAAAGATCC-3´, which were described by Noda et al and amplify a 146 bp region of DNA unique to pathogenic Leptospira spp., the lipL32 gene.14 To do this, 2.5 µL of PCR buffer (10X), 0.75 µL MgCl2 (50 mM), 0.5 µL dNTP's (10 mM), 0.75 µL of each primer (10 µM), 0.25 µL Taq Polymerase (Taq DNA Polymerase Recombinant from Invitrogen), 15.5 µL of molecular grade water, and 3 µL of DNA were used for a final reaction of 24 µL.
Leptospira interrogans DNA was used as a positive control and sterile water as a negative control. The amplification profile by Tique et al was proposed with some modifications: 1 cycle at 95°C for 3 minutes, followed by 35 cycles at 95°C for 45 seconds, 60°C for 30 seconds, and 72°C for 30 seconds.15 The PCR products were analyzed using 1.5% agarose gel and visualized using SYBR safe on a UVP transilluminator.
The amplified products were purified with a commercial kit (PureLinkTM Quick gel Extraction), following the manufacturer's recommendations. The samples were sent to Macrogen Inc (Seoul, South Korea) for sequencing. The resulting electropherograms were edited and aligned using MEGA-X software to obtain a consensus sequence per sample. These, in turn, were aligned with sequences recorded in GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using the Basic Local Alignment Search Nucleotide (BlastN) tool from the National Institutes of Health (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to determine and compare their identity and similarity.
Ethics statement
The procedures for capture, manipulation, euthanasia, and identification of the biological material were approved by the Ethics Committee of the Universidad Cooperativa de Colombia by virtue of ethical concept No. 029-2017. This project had the authorization of the National Authority of Environmental Licenses (ANLA, by its Spanish acronym) by the Universidad de Córdoba, within the framework of the collection of biological samples resolution 0914 of August 4, 2017, regulating the collection of specimens of wild species from biological diversity for non-commercial biodiversity research, in accordance with the provisions contained in Decree 1376 of 2013, now compiled in Decree 1076 of 2015.
Results
50 rodents from the genera Rattus rattus 30 (60%), Mus musculus 8 (16%), Zygodontomys brevicauda 8 (16%), Oligoryzomys sp. 2 (4%), Hylaeamys (formerly Oryzomys) 1 (2%), and Proechimys cf. oconnelli 1 (2%) were collected across the 7 townships belonging to the municipality of Villavicencio.
Detection of molecular markers for pathogenic leptospira
Of the 50 kidney tissue samples, 12% (6/50) were positive for the markers of the lipL32 gene found in most species of pathogenic leptospira, amplifying a product of 146 bp. Four of these rodents belonged to the species Rattus rattus, 1 to Zygodontomys brevicauda and 1 to Oligoryzomys sp. (Figure 1).
A total of 66.7% (4/6) of captures were made in open field areas and 33.3% (2/6) within peridomiciliary areas. The incidence of infection based on species was 8% (4/50) for Rattus rattus, 2% (1/50) for Zygodontomys brevicauda, and 2% (1/50) for Oligoryzomys sp. (Figure 2). The incidence of infection based on the sex and capture site of each species can be seen in (Table 1).
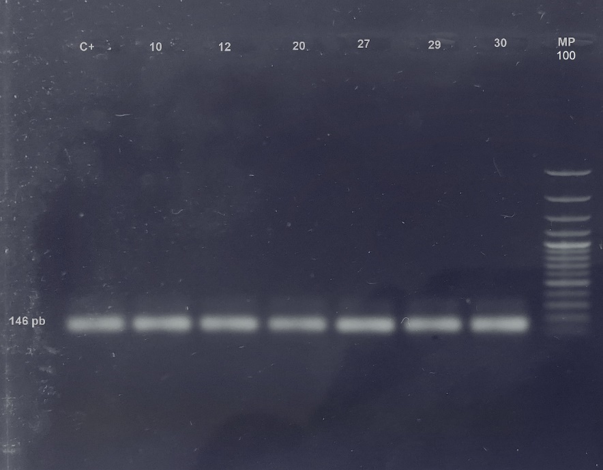
Fig 1 - Amplification of lipL32 gene from Leptospira interrogans. 1.5% agarose gel with SYBR safe. An amplification of 146 bp can be observed. C+: Positive control; 10, 12, 29 and 30: Rattus rattus; 20:Zygodontomys brevicauda; 27: Oligoryzomys sp.; MP: Molecular weight marker.
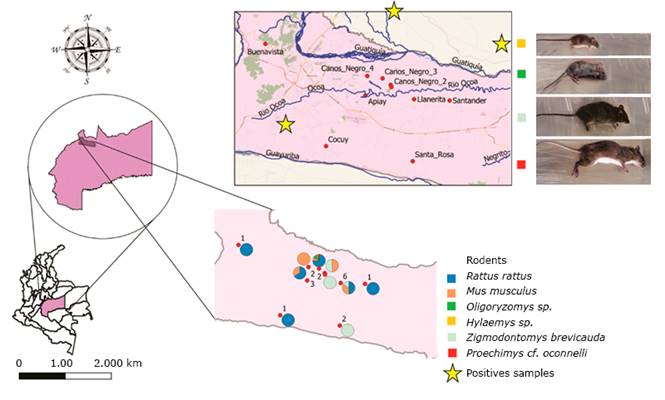
Fig 2 -Geographical area, distribution, and identification of rodents captured in the municipality of Villavicencio, which were positive for Leptospira spp.
Table 1 -Area and identification of small rodents with a positive PCR for pathogenic leptospira
| % | |||||
|---|---|---|---|---|---|
| Cocuy | Peridomiciliary | Fa | 2 | ||
| Santander | Peridomiciliary | Mb | 2 | ||
| Caños negros | Countryside | M | 8 | ||
| Countryside | M | ||||
| Countryside | M | ||||
| Countryside | M | ||||
| Total | 12 | ||||
Sequencing and genetic analysis
A total of 6 consensus sequences were obtained from the kidney samples of Rattus rattus, Zygodontomys brevicauda, and Oligoryzomys sp in the peri-urban areas and the countryside of the municipality of Villavicencio. The alignment of these sequences revealed that 6 of them showed greater homology with Leptospira interrogans, and that the first nucleotide was homologous to the nucleotide located at position 545 of the lipL-32 gene of L. interrogans consigned in GenBank under access number MN373267.1. The alignment analysis carried out using the BlastN tool yielded coverage and identities of 98.64% with respect to the pathogenic species Leptospira interrogans, which were close to the serovars Hardjo and Canicola (Table 2).
Discussion
The presence of pathogenic leptospira in the kidney of synanthropic and wild rodents is a significant finding as there are no previous reports of infection in these rodents in the areas under the municipality of Villavicencio. Previous studies have established the importance of leptospirosis as the second cause of undifferentiated febrile illness in this municipality after dengue.
Rodents are common carriers and disseminators of Leptospira. A wide variety of rodent’s species in almost all regions in the world are chronic carriers of these bacteria; however, these mammals do not present any noticeable clinical distress. The presence of leptospira in the kidney of these rodents entails frequent emissions of the bacteria through their urine for prolonged periods.16
Of the 50 rodents captured in the rural and peri-urban areas of the municipality of Villavicencio in this study, 12% were infected with pathogenic leptospira, which is a high percentage when compared with that reported in the study conducted by Torres et al in Yucatán, Mexico.17 In that study, of the 92 captures of synanthropic and wild rodents, 5.4% were positive for leptospirosis. It should be highlighted that, in the study by Torres, of the 5 rodents infected with leptospira, 2 belonged to the Rattus rattus species and 2 belonged to Mus musculus. Conversely, in our study, 4 of the 6 rodents were R. rattus and 2 were wild species. The importance of these wild species is that they can become maintenance hosts depending on the infectious serovar and on environmental factors and may pose a potential risk for the human populations with which they come into contact.18
For a long time, most studies on leptospirosis had focused on the species Rattus rattus and Rattus norvegicus. In pig farms from the coffee region, Giraldo et al. found that of 75 captures of rodents, 62 (82.7%) showed evidence of infection by leptospira.19 In addition, those studies proved the presence of the serovars Javanica, Australis, Bataviae, Canicola, and Pomona among others. However, more recent studies have identified various genera and species of wild rodents as reservoirs of Leptospira spp. Torres et al. in Yucatán, Mexico, captured 47.8% of wild rodents, although only one Heteromys gaumeri was positive for Leptospira interrogans.17
The reproductive capacity of rodents makes them pests that contaminate water sources for human and animal consumption as they live in their vicinity and can easily access facilities, being found in drains, sewers, garbage dumps, walls and ceilings. Additionally, rodents can be found in food storage sites and in animal feeders in a higher proportion. They contaminate these places with their urine rather than by eating the concentrates, which favors the transmission and dissemination of spirochetes. Rodents are maintenance hosts mainly for the serovars Icterohaemorrhagiae and Copenhageni.20 Regarding Hardjo and Canicola serovars, which yielded close BlastN values in this study, it can be considered that the Hardjo serovar is the most prevalent serovar within bovine cattle. Thus, contaminated water and open herds may be factors that favor interspecies cross contamination and dissemination in rodents.
The Canicola serovar has been associated with dogs, and it is important to consider that dogs are susceptible to infection given their high exposure to the pathogen, even in urban areas. The main sources of leptospira are irrigation and drainage water, as there is continuous elimination of the bacteria through the urine by rodents and other animals.21
A study conducted by Sánchez et al. that included the municipality of Villavicencio detected seroconversion in paired serum samples from patients with undifferentiated febrile illness using the MAT technique in 29% of the samples.12 A total of 37.93% of these samples belonged to the Canicola serovar, which is consistent with the serovar found in our study. Despite the small number of captures in our study, the results could be an indication of the role of rodents, dogs, and cattle as contaminants of stagnant water and water for consumption in the epidemiology of this disease in the municipality of Villavicencio.
The list of the best results from the BLAST analysis revealed the presence of the lipL32 gene in the 6 pathogenic strains found in this study. lipL32 is a highly preserved gene among the pathogenic species of leptospira and is even completely absent in the saprophytic species of L. biflexa. Molecular techniques, such as PCR have proven effective in the identification of leptospira as they are fast, sensitive, and specific and have become a useful tool in epidemiological surveillance, establishing the basis for control and prevention programs in our region.
In conclusion, the rodents captured at the study site were indeed reservoirs of pathogenic leptospira and could be part of the infection cycle of leptospirosis in the region. To the best of our knowledge, this is the first molecular evidence of the circulation of Leptospira interrogans sensu lato in wild and synanthropic rodents in the municipality of Villavicencio.














