Introduction
Emerging viruses are becoming a great danger to global health and among them, coronaviruses are a notable example. Very virulent forms have emerged from their natural animal hosts and represent a threat to human communities. Between 2002-2003, Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV)1,2 arose in China from bat populations, passing to civets and finally to humans. Ten years later, Middle East Coronavirus Respiratory Syndrome Coronavirus (MERS-CoV)3 also arose from bats, transferring in the Middle East to dromedary camels and then to humans. Recently, at the end of 2019, another virus emerged in Wuhan, Hubei Province, China, which has been recognized as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2),4,5 responsible for coronavirus disease 2019 (COVID-19).
Current coronavirus classification recognizes 39 species in 27 subgenus, five genus and two subfamilies that belong to the Coronaviridae family, Cornidovirineae suborder, Nidovirales order and Riboviria realm. Within the Coronaviridae family is the Orthocoronavirinae subfamily, which is made up of four genera, which according to their genetic structure are grouped into taxa called Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus. Both alphacoronaviruses and betacoronaviruses cause different diseases in different mammalian species,6 including respiratory infections and gastroenteritis. SARS-CoV-2 virus belongs to the severe acute respiratory syndrome related coronavirus species among the group of betacoronaviruses.7
Many aspects of the structure and biology of the SARS-CoV-2 virus have not yet been elucidated. Development of effective preventive and therapeutic strategies may be hindered by the lack (need) of information related to the structural details of viral proteins, although some crystallographic structures8,9) are already available. This review describes the general aspects of the structure of the SARS-CoV-2 virus, referring to the characteristics of the proteins encoded by the viral genome.
Methods
A review was carried out between March and May 2020. 47 bibliographic references were analyzed, which correspond to articles from national and international journals available in the PubMed, Scopus, Medline and SciELO databases. The most recent published literature in relation to the subject area studied was considered as the bibliography selection criterion. Web pages of the Ministerio de Salud Pública de Cuba, World Health Organization and Pan-American Health Organization were explored. For the collection of information, a search strategy was applied using health science keywords and connectors, and analysis-synthesis and logical deduction methods were applied to write the article.
General structure of SARS-CoV-2
Coronaviruses are enveloped viruses with a single-stranded, positive-sense RNA molecule.10 They belong to a large family of viruses that infect birds and various mammals, including camelids, bats, civets, rats, mice, dogs, and cats. Viruses in this family are 60-220 nm size and filamentous forms from 9-13 nm in diameter are usually observed.11
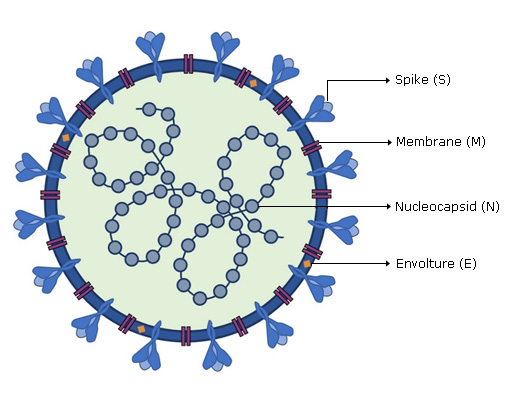
SARS-CoV-2 viral genome encodes at least sixteen non-structural proteins (nsps) and four structural proteins. Structural proteins (Fig. 1) form the viral particle that consists of a nucleocapsid, formed by the viral genome to which multiple copies of nucleocapsid (N) protein are attached, which is surrounded by an envelope where proteins are inserted: spike (S), membrane (M) and envelope (E) proteins are inserted.
SARS-CoV-2 Genome organization
The virus has a positive-polarity single-stranded RNA genome, 26-32 kb long.12 From this molecule, proteins that are necessary to finish the complete replication cycle are synthesized. These proteins include a replicase transcriptase complex that produces more RNA and various structural proteins that build new virions.
SARS-CoV-2 genome is similar to that of the SARS-CoV and MERS-CoV viruses, with 88 % and 50% of sequence identity,13 respectively. In 5'-3' sense, the genome is organized into fourteen open reading frames (ORFs) (Fig. 2a) that encode a variety of structural or non-structural proteins, according to their functions in the viral particle.
 A- Schematic diagram of the sequence of ORFs in viral RNA. B- Schematic diagram of the pp1a and pp1ab polyproteins encoded by the ORF1a and ORF1b. PLpro, papain-like proteases; TM, transmembrane; Mpro, chymotrypsin-like protease; RdRp, RNA-dependent RNA polymerase; Hel, helicase; Exo, 3 '→ 5' exonuclease; NU, NendoU: uridylate-specific endoribonuclease; MT, ribose-2′-O-methyltransferase; nsp, non-structural protein. Created for this article.
A- Schematic diagram of the sequence of ORFs in viral RNA. B- Schematic diagram of the pp1a and pp1ab polyproteins encoded by the ORF1a and ORF1b. PLpro, papain-like proteases; TM, transmembrane; Mpro, chymotrypsin-like protease; RdRp, RNA-dependent RNA polymerase; Hel, helicase; Exo, 3 '→ 5' exonuclease; NU, NendoU: uridylate-specific endoribonuclease; MT, ribose-2′-O-methyltransferase; nsp, non-structural protein. Created for this article.Fig. 2- Structure of the SARS-CoV-2 virus genome.
During the transformation phase, the ORF1a and ORF1b sequences, which correspond to almost two thirds of the virus RNA, are translated into two large overlapping polyproteins called pp1a and pp1ab,14 which are processed into sixteen non-structural proteins (nsps 1-16), many of which form the replicase transcriptase complex.15 The nsps 3 and 5 supplement the enzymes papain-like proteases (PLpro)16) and chymotrypsin-like protease Mpro (3CLpro),17 respectively, whom decides the processing of viral polyproteins. Cleavage occurs between the products of ORF1a and ORF1b to form pp1a made up of nsps 1-11, while pp1ab will be made up of nsps 1-16 (Fig. 2b). The other SARS-CoV-2 ORFs that correspond to one third of the viral genome, encode for the four structural proteins N, S, M and E, as well as other accessory proteins whose function is not yet known, although it is known that they don’t participate in viral replication in cultured cells.
SARS-CoV-2 non-structural proteins
SARS-CoV-2 non-structural proteins have different functions that decide several processes in the virus and in the host cell. The non-structural protein (nsp) 1 promote inhibition of type 1 interferon (IFN) signaling and block the innate immune response in the host cell by degrading the host mRNA, inhibiting translation and stopping the cell cycle,18 while nsp2 binds to a proinhibitory protein whose function is still unknown. In addition to guaranteeing the processing of the pp1a and pp1ab polyproteins, nsp3 and nsp5 promote the expression of cytokines.19 The nsps 4 and 6 contribute to the structure of the double membrane vesicles by determining the transmembrane protein framework.20,21 nsp7/8 complex is a hexadecameric complex that support of the replication enzyme,22 where nsp8 constitutes a second RNA polymerase that can function as a primase. RNA-dependent RNA polymerase is represented by nsp12, which together with the helicase RNA (nsp13) guarantee the assembly of the replicase transcriptase complex.23 nsp9 constitutes an RNA-binding protein phosphatase, which like nsp10, is part of the replicase complex. nsp10 establish also a necessary link for the adequate function of the main viral protease (Mpro).19 nsp14 is a bifunctional protein, which exhibits exonuclease activity 3'→5' with a role in maintaining the fidelity of RNA transcription,24 and (guanine-N7)-methyl transferase activity, involved in RNA cap formation.25 RNA cap formation is also facilitated by nsp16, which is an S-adenosyl-methionine RNA-dependent with (ribose-2'O)-methyl transferase activity.26 Finally, nsp15 encode a uridylate-specific endoribonuclease (NendoU)27 that is crucial for virus replication and distinguishes nidoviruses from other RNA viruses. These and other functions are summarized in table.
Table- Functions of the non-structural proteins of the SARS-CoV-2 virus
| Non-structural proteins | Function |
|---|---|
| nsp1 | Inhibition of type 1 IFN signaling; block of the innate immune response in the host cell. |
| nsp2 | Binds to a proinhibitory protein of unknown function. |
| nsp3 | PLpro activity for processing the pp1a and pp1ab polyproteins; cytokine expression; IFN-β antagonist; deubiquitinase activity. |
| nsp4 | Double membrane vesicles formation. |
| nsp5 | 3CLpro activity for processing of pp1a and pp1ab polyproteins; cytokine expression. |
| nsp6 | Double membrane vesicles formation. |
| nsp7 | Hexadecameric complex formation. |
| nsp8 | Primase; hexadecameric complex formation. |
| nsp9 | RNA binding protein phosphatase. |
| nsp10 | Part of the replicase complex. |
| nsp11 | Unknown. |
| nsp12 | RNA-dependent RNA polymerase. |
| nsp13 | RNA Helicase. |
| nsp14 | 3'→5' exonuclease activity; N7-MTase activity for the RNA cap formation. |
| nsp15 | Uridylate specific endoribonuclease. |
| nsp16 | 2'O-MTase activity for the RNA cap formation. |
PLpro, papain-like proteases; IFN, Interferon; 3CLpro, chymotrypsin-like protease Mpro; N7-MTase, (guanine-N7)-methyltransferase; 2'O-MTase, nucleoside-2'O-methyl transferase.
The three-dimensional structure of the main SARS-CoV-2 protease is very similar to that of SARS-CoV, sharing a sequence identity of 96 %.8 This enzyme is essential to translated polyproteins of viral RNA. Mpro operates at no less than eleven cleavage sites on the ORF1ab polyprotein. The recognition sequence in most of these sites is Leu-Gln, while the Ser, Ala, Gly residues mark the cleavage site.17
Mpro is a dimer of two identical subunits that together form two active sites,28 each of which have three domains identified as I, II and III. This dimerization of the enzyme is necessary for its catalytic activity. At the catalytic site Cys145 and His41 amino acids carry out the protein cleavage reaction. In contrast to SARS-CoV virus, SARS-CoV-2 protease presents the substitution of Ser284, Thr285 and Ile286 by Ala residues which leads to 3.6 times progress of the catalytic activity of the protease, which go together with a lightly closer packaging of both III domains of each subunit, leading to greater stability.8,17
SARS-CoV-2 structural proteins
M protein is the most abundant structural protein, which is responsible for shaping the virion. Multiple sequence alignment points show remarkable similarity between the Sars-CoV-2 sequences and those isolated from Pangolin-CoV-MP798 and Bat-CoV-CoVZXC21.29,30 However, there is heterogeneity at the N-terminal, where an insert of a Ser residue at position 4 of SARS-CoV-2 M protein appears to be a unique feature.31
M monomer ranges from 25 to 30 kDa, and it is embedded in the envelope through three transmembrane domains.32 N-terminal constitutes a small ectodomain, while the C-terminal endodomain is located on the inner face of the virion membrane contributing to most of the molecule. The ectodomain can be modified by glycosylation, which influences the tropism of the organs to be infected. This protein is responsible for the transmembrane transport of nutrients, the virion release and the envelope formation. Binding with the M protein helps stabilize the N proteins and promotes the termination of the viral assembly by stabilizing the RNA-N protein complex within the internal virion.33
N protein forms the helicoidal nucleocapsid, joining along the entire viral genome, which assumes a curl shape. This protein weighs 43-50 KDa and is phosphorylated in a discrete number of serines and threonines. Although the role of this phosphorylation has not yet been determined, it has been suggested that it is related to regulatory functions and its ability to bind to the viral genome.33
N protein contains two domains that allow it to recognize viral RNA. It is also capable of binding to nsp334 to address the genome to the replicase transcriptase complex and ensure nucleocapsid packaging. It also works as an IFN-β antagonist,35 contributing to the mechanisms of viral evasion to the immune system, by preventing interferon from stopping viral replication in still healthy cells and the destruction of already infected cells, highlighting the importance of exogenous administration of interferon in the treatment of disease caused by the virus. N protein also participates in the repression of host cell interference RNAs that suppress the expression of specific sequences of the viral genome and that constitute a vital part of the body's immune response to viruses.36
E protein is a small polypeptide that is found in limited amounts in the viral envelope. In particular, the E protein sequence of SARS-CoV-2 is identical to that isolated from Pangolin-CoV-MP798 and from Bat-CoV-CoVZXC21, CoVZC45 and RaTG13.32 Two distinguishing features of Sars-CoV-2 E protein when compared to that of other homologs is the substitution at position 69 where an Arg replaces Glu, Gln, Asp, and deletion at position 70 corresponding to Gly or Cys in another E proteins.37
During the replication cycle, this protein is abundantly expressed within the infected cell, but only a small amount is incorporated into the envelope of the virion. Most of it is located in the intracellular traffic site, such as the Golgi complex, where it participates in the assembly of the particle and it is very important in the production and maturation of the viral particle.37
SARS CoV-2 virus glycoprotein S is a densely glycosylated trimer that projects in the form of spicules and it is 16-21 nm long.38 It is a typical class I viral fusion protein that requires the cleavage of a protease for the activation of its fusion potential, so that during viral infection,39 it is cleaved by a furin-like cellular protease into two peptides of the same size, S1 and S2. There is a cleavage site between the S1 and S2 subunits (amino acids 682-685, RRAR) that creates a polybasic furin site that has been linked to rapid virus transmission, by facilitating endocytosis mechanisms to human cells (Fig 3 a ).40
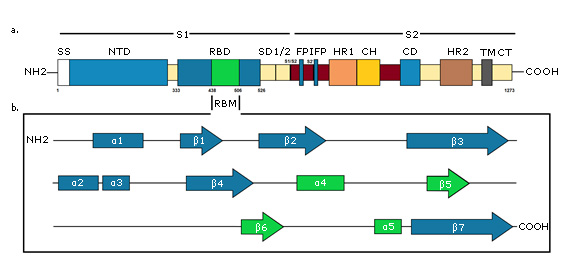 A - Primary structure of S protein. SS, signal sequence; NTD, N-terminal domain; RBD, receptor-binding domain, RBM, receptor-binding motif; SD1/2, subdomain 1 & 2; S1/S2 and S2', protease cleavage site; FP, fusion peptide; IFP, internal fusion peptide; HR1, heptad repeat 1; CH, central helix; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. B - Secondary structure of S protein RBD. RBM is green colored. Created for this article.
A - Primary structure of S protein. SS, signal sequence; NTD, N-terminal domain; RBD, receptor-binding domain, RBM, receptor-binding motif; SD1/2, subdomain 1 & 2; S1/S2 and S2', protease cleavage site; FP, fusion peptide; IFP, internal fusion peptide; HR1, heptad repeat 1; CH, central helix; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. B - Secondary structure of S protein RBD. RBM is green colored. Created for this article.Fig. 3- Schematic diagram of the SARS-CoV-2 S protein.
S1 subunit is highly variable between different coronaviruses. In the monomeric structure of S protein, N and C-terminal portions of this subunit fold as two independent domains, the N-terminal domain (called NTD) and the C-terminal domain (or C domain).38 Depending on the virus, NTD or the C domain can serve as the receptor binding domain (RBD). In SARS-CoV-2, the S1 C domain has the RBD of approximately 21 kDa and 200 amino acid residues.9 The RBD subdomain is responsible for S protein organizing in the trimer form and for binding directly to the peptidase domain (PD) of Angiotensin-Converting Enzyme 2 (ACE2) of human cells.41,42 In contrast, S2 is highly conserved and the fusion peptide is located in it, which is responsible for the fusion of the viral membrane to the host cell membrane, as well as the cytopathic effect that this virus can produce when it infects cells in vivo.38
The trimeric structure of the SARS-CoV-2 S protein has one RBD in one up conformation and two RBD in down conformations. This process is triggered when the S1 subunit binds to the host cell receptor.43 Receptor binding destabilizes the prefusion trimer, resulting in detachment of the S1 subunit and transition of the S2 subunit to a stable post-fusion conformation. To compromise a host cell receptor, S1 RBD undergoes hinge-like conformational movements that temporarily hide or expose the determinants of receptor binding.44 These two states are referred to as down conformation and up conformation, where the first corresponds to the inaccessible receptor state, and the second corresponds to the accessible receptor state, which is stable. Observation of this phenomenon in the SARS-CoV-2 S protein suggests that it shares the same activation mechanism that is believed to be conserved in the Coronaviridae family, in which binding of the receptor to exposed RBDs leads to elimination of S1 and the folding of S2.
The final RBD model of the SARS-CoV-2 S protein contains residues from Thr333 to Gly526. This subdomain has a twisted five-stranded antiparallel β sheet (β1, β2, β3, β4 and β7) with short helices and connection loops forming the core. Between the β4 and β7 strands in the core, there is an extended insertion containing short β5 and β6 strands, α4 and α5 helices and loops (Figure 3b). This extended insertion is the receptor binding motif (RBM) that contains the majority of the SARS-CoV-2 contact residues for ACE2 binding. There are nine cysteine residues in the RBD, eight of which form four pairs of disulfide bonds. Among these four pairs, three are in the core (Cys336-Cys361, Cys379-Cys432, and Cys391-Cys525) to help stabilize the structure of the β sheet structure, while the remaining (Cys480-Cys488) connects the loops at the distal end of the RBM.44,45
The presence of RBD in the virus S protein determines that it plays the most important roles in viral binding, fusion and entry into the host cell, making it a promising target for the development of antibodies, entry inhibitors and vaccines for the prevention and treatment of the disease46,47.