Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista de Protección Vegetal
versión impresa ISSN 1010-2752
Rev. Protección Veg. vol.29 no.3 La Habana sep.-dic. 2014
ORIGINAL ARTICLE
Molecular detection of Potato yellow vein virus in the natural whitefly vector Trialeurodes vaporariorum, Westwood
Detección molecular de Potato yellow vein virus en el vector natural Trialeurodes vaporariorum, Westwood (mosca blanca)
Carlos Eduardo Barragan, Mónica Guzmán-Barney*1
Instituto de Biotecnología, Universidad Nacional de Colombia, Carrera 30 #45-30, Bogotá DC, Colombia. Código postal 111321. 1Plant Viruses Laboratory Coordinator.
ABSTRACT
Potato yellow vein virus (PYVV) expresses yellowing symptoms on plants and it is transmitted by the whitefly vector Trialeurodes vaporariorum Westwood. No molecular viral detection from the vector extracts has been reported to date. The objectives of this study were I) amplification of the PYVV major coat protein (CP) gene from viruliferous vector extracts by RT-PCR using specific primers, II) cloning and III) sequencing and sequences analysis. Some Colombian Solanum phureja (Jus et Buk) field potato plants expressing yellowing symptoms and confirmed PYVV positive by RT-PCR were used as virus donor plants for feeding whiteflies. Viruliferous vectors were obtained by depositing aviruliferous whiteflies on symptomatic leaves and kept in a muslin cloth-covered glass flask for 48 h. A kit SV for total RNA Isolation System (Promega®) was used for extracting the viral RNA from one viruliferous whitefly and from pools of viruliferous vectors (2, 5, 10, 15 and 20 individuals); CP gene amplicons of about 765bp were cloned and sequenced to confirm the PYVV genome. Blast alignment established more than 99% similarity between CP gene amplified from vector extracts and several GenBank reported PYVV CP genes from Colombian and Peruvian isolates.This study presents information concerning the molecular detection of PYVV in its natural vector for the first time by using a very simple viral acquisition technique for obtaining viruliferous vectors that could be of interest for future studies concerning virus and vector gene characterization, plant-virus and vector interaction and/or epidemiological studies.
Key words: PYVV, RNA, whitefly, detection, vector.
RESUMEN
El Potato yellow vein virus (PYVV) produce síntomas de amarillamiento en las nervaduras de la hoja de papa, y es transmitido por el vector natural Trialeurodes vaporariorum Westwood (mosca blanca). Hasta la fecha no se ha detectado molecularmente PYVV en extractos de moscas virulíferas. Los objetivos de este estudio fueron I) amplificar el gen de la proteína mayor de la cápside (CP) de PYVV a partir de extractos del vector, utilizando RT-PCR con cebadores específicos, II) clonar los amplicones y III) secuenciar y analizar las secuencias. Se utilizaron como donadoras de virus, las plantas de papa Solanum phureja (Jus et Buk), colectadas en campo con síntomas de amarillamiento y confirmadas positivas para PYVV por RT-PCR. Se depositaron moscas aviruliferas en hojas sintomáticas cubiertas con tela de muselina, para la adquisición de PYVV durante 48 h. Para la extracción de RNA viral se empleó el kit SV total RNA Isolation System (Promega®), utilizando una mosca o grupos (2, 5, 10, 15 y 20 individuos). Los amplicones obtenidos de CP, aproximadamente 765 pb se clonaron y secuenciaron. Las secuencias obtenidas se alinearon con la herramienta BLAST; se establecieron similitudes del 99% entre el amplicón derivado del vector y secuencias del gen CP de aislados de PYVV colombianos y peruanos.
Palabras clave: PYVV, RNA, mosca blanca, detección, vector.
INTRODUCTION
Potato yellow vein virus (PYVV) is a re-emergent pathogen affecting Solanum phureja (Jus et Buk) and other potato plants in Andean countries. PYVV requires quarantine measures being taken as it causes yellow vein disease (PYVD) from the leaf apex until the leaflets become totally chlorotic (1, 2). Yields of infected plants reduced by up to 50% in plots where all the plants had become infected (2) with reports of up to 30% in Solanum phureja (3, 4).
PYVV is a member of the Closteroviridae family, genus Crinivirus (5), having flexuous filamentous particles which are limited to the plant phloem (2, 6, 7). The genome is tripartite, consisting of 8 kb, 5.3 kb and 3.8 kb RNA positive-sense, single-stranded segments (RNA1, RNA 2 and RNA 3, respectively); two defective RNA (dRNA) have also been reported (8). RNA 1 contains the replication module and RNA 2 and 3 contain the hallmark Closteroviridae gene array which includes the major coat protein (CP) and the heat shock protein 70 homolog(Hsp70h) (8).
So far, the molecular detection and characterisation of PYVV have been reported from leaf extracts (4, 9, 10, 11, 12, 13). PYVV is transmitted by infected tubers (14) and semi-persistently by the natural whitefly vector (Trialeurodes vaporariorum, Westwood) (2). High PYVD incidence in Colombia (3, 14) is due to increased vector populations, although indiscriminate use of potato-infected seed provides the major pathway for viral dissemination (2).
Whitefly-transmitted viruses have become increasingly prevalent in crops grown in tropical and subtropical parts of the world (15, 16), thereby leading to the emergence of numerous whitefly-transmitted diseases (17, 18, 19). The whitefly Bemisia tabaci has been reported as a vector of viruses such as Tomato yellow leaf curl virus (TYLCV), Tomato infectious chlorosis virus (TICV), and Tomato chlorosis virus (ToCV) (20, 21, 22). On the other hand, Cucumber vein yellowing virus (CVYV) and Cucurbit yellow stunting disorder virus (CYSDV) have been detected by RT-PCR from B. Tabaci extracts (23, 24).The interaction between T. vaporariorum, PYVV and a host (potato/tomato) has only been reported by classical vector transmission and symptom expression assays(25).
No reports concerning the molecular detection of PYVV from T. vaporariorum extracts have been reported to date.The aim of this study was to detect PYVV from viruliferous T.vaporariorum extracts for the first time by using molecular methods and using a short procedure to obtain viruliferous whiteflies.
MATERIALS AND METHODS
Virus isolates
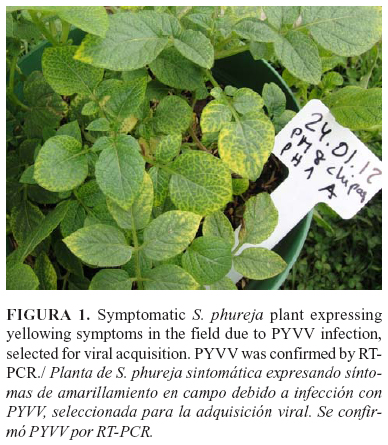
Two Colombian Solanum phureja potato plants showing yellowing symptoms were harvested in a field near Chipaque, Cundinamarca, Colombia. The tubers from these plants were planted and shoots expressing yellowing symptoms (Fig. 1) were diagnosed for PYVV by RT-PCR. The viral RNA was extracted from the petioles; PYVVCP and the Hsp70h genes were amplified using previously-described specific primers (11, 26).
PYVV acquisition by the insect vector
Aviruliferous vector: A nursery of whiteflies (T. Vaporariorum Westwood) was obtained by feeding them on PYVV-resistant green zucchini plants grown in a greenhouse (15°C to 25°C).
Viruliferous vector: A large population of aviruliferous whiteflies (100) was divided into samples containing 1 and 2 whiteflies and pools of 5, 10 and 20 whiteflies (with duplicates). Each whitefly or pools were left to feed on a single symptomatic potato leaf (Fig. 1) kept in a distilled water-filled beaker. The flask was covered with a film and a muslin veil cage was attached to the underside of the leaf (Fig. 2). The vector was allowed to feed on the virus-infected leave for 48 h for the acquisition access periods (AAP). Aviruliferous whiteflies were placed to feed on symptomless potato leaves as controls (negative for PYVV by RT-PCR).
PYVV detection from the vector: The viruliferous whiteflies were quickly immobilised by briefly chilling at 4°C and the cages were carefully removed from the potato leaves. The 1, 2, 5, 10, 15 and 20 whitefly samples were picked up and used for extracting the vector total RNA.
RNA extraction from whiteflies
Total RNA was extracted from each whitefly sample or pool using an SV total RNA Isolation System,Promega®, Madison, USA.The whiteflies were carefully picked up from the leaves using a wet needle, directly placed into microtubes containing the buffer, and ground using a plastic pestle. RNA concentration was determined following extraction using a Qubit® spectrophotometer (Invitrogen®).
RT-PCR and primer design
All RT-PCR amplifications were performed using F2oligonucleotide primers
(5'-GGATCCTCATGGAAATCCGATC-3') and (5'-CTACTCAATAGATCCTGCTA-3') for amplifying an extended PYVV CP gene region (13); the Hsp70h gene was partially amplified with F517 (5'-AGAGACGGTAAGTAT-3') and R2323 primers (5'-TTGGGCATGTGACAT-3') designed by Cubillos (26) from a Peruvian isolate genome sequence (NC-006063.1). Trialeurodes vaporariorum control primers Tv18SR (5'-CGTTCGGGGTTTATCTTTATC-3') and Tv18SF(5'-CAGACCGTGACTTTTGAAC-3') were designed using the 18S region of its rRNA as a template (accession No. HQ446161.1). These primers were thoroughly tested for primer-dimers using OligoAnalyzer 3.1 software (ADT) and no similarity to any plant or viral RNA. All the primers were provided by ADTTechnologies®.
CP and Hsp70 gene amplification:
cDNA synthesis involved whitefly RNA extracts (10 ml) being heated at 70°C for 5 min, rapidly chilled on ice and then added to the RT-PCR mix containing 0.4 mM virus-specific reverse primer, 3.2 U broad-spectrum RNase inhibitor(RNasin,Fermentas®), 1XRT buffer (Epicentre®), 1 mM dNTPs, 10 mM dithiothreitol and 16U Moloney murine leukaemia virus (MMLV) reverse transcriptase (Epicentre®), in 10 ml reaction volume. RT-PCR was run at 42°C for 45 min in a thermal cycler. cDNA from PYVV-infected S.phureja symptomatic leaves and healthy S.phureja leaves was used as a positive control and cDNA from aviruliferous whiteflies as a negative control; water was included as a blank reaction.
A volume of 10 ml was used for the PCR reaction, containing 5 ml cDNA, 1XPCR buffer (Bioline®), 250 mM dNTPs, 0.4 mM of each primer and 0.8 U Biolase® DNA polymerase (Bioline®). Amplification involved denaturing at 94°C for 3 min followed by 30 cycles at 94°C for 30 s, 60°C for 30 s and 72°C for 60 s. The samples were incubated for an additional 10 min at 72°C. Each RT-PCR reaction was run in triplicate, with 5 to 8 replications per transmission. PCR products were migrated onto stained 1% agarose gel using TAE buffer with SybrSafe (Invitrogen®).
The primers designed for amplifying the vector rRNA 18s region were tested in different reactions and in the same RT-PCR reaction in which the CP and Hsp70h genes were amplified to confirm an appropriate RNA extraction from T. Vaporariorum and avoid false negatives due to the presence of PCR inhibitors.
Amplicon sequence and bioinformatics analysis
The CP amplicons were ligated into TOPO-TA vector (Invitrogen), according to the manufacturer's instructions, and transformed into Escherichia coli-TOP10 cells to confirm amplification of the T. vaporariorum-derived PYVV CP gene. The plasmids were extracted from the selected colonies using a Qiagen Plasmid QuickLyse® kit and linearised by digesting with EcoRI (Promega®). Both strands were sequenced using Universidad Nacional de Colombia's sequencing laboratory SSiGMol amplification primers (Applied Biosystems® 3500 Series Genetic Analyzer).
Electropherogram results were assembled using BioEdit software and debugged sequences were used for predicting the protein using EMBOSS Sixpack software. Blast was then used for comparing this with the already published sequence information.To confirm sequence identity and predicted CP, sequences were aligned in Muscle (27) and used for a phylogenetic reconstruction at http://www.phylogeny.fr (28). The results were analysed using the neighbor-joining method (1,000 bootstraps, Jones-Taylor-Thornton substitution matrixmodel) (29,30). The GenBank Peruvian PYVV YP-054421 sequence was used as a control for the cladogram which also included the AEC22851.1 sequence from a PYVV isolated from Colombian S. phureja. The CP gene sequence reported for Citrus tristeza virus (CTV) (accesion code NP-042867.1) was used for the base of the cladogram; both CTV and PYVV belong to the Closteroviridae family.
RESULTS
RT-PCR detection of PYVV in viruliferous whiteflies
For the first time, it was obtained viruliferous vectors (capable of transmitting virus in a semipersistent way) using one symptomatic plant leaf placed in a small glass flask containing distilled water and covered with a muslin cage.
The RNA extracted from whiteflies using the aforementioned kit ranged from 12 ng/ml (1 whitefly) to 46 ng/ml (20 whiteflies), thereby ensuring the success in further CP gene and Hsp70h gene amplifications. CP gene and HSP70h amplicons were estimated at 765bp and 1,656bp, respectively (Fig. 3a-b). No positive RT-PCR results were obtained from the aviruliferous whiteflies feeding on healthy S. Phureja plants.
The 18S rRNA region of the vector genome was amplified from the RNA extracts to guarantee the absence of PCR inhibitors in the vector extract and avoid false negatives. The expected band of around 290 bp for the 18SrRNA region was obtained from both viruliferous and aviruliferous whiteflies feeding on zucchini (Fig. 3b).This approach was carried out many times (for one or insect pools) giving consistently positive results for viral genes amplification by RT-PCR.
The electrophoresis gel (Fig. 3a) revealed a CP gene migration amplicon of around 790 bp, with 20 bp more than the expected PYVV CP size (760 bp) because the forward F2 primer annealed in an upstream region of the CP gene. The PYVV Hsp70h gene was also successfully amplified.
Nucleotic sequences and bioinformatics analysis
The CP gene sequences obtained were compared with those reported in GenBank. The sequence analysis and alignment revealed >99% similarity with CP genes of different Colombian PYVV isolates from extracts obtained from symptomatic leaves (different accessions reported in GenBank), and 98-99% similarity with a Peruvian PYVV isolate (YP-054421). Sequences obtained in this study were submitted to GenBank (accession numbers KC257442, KC257443, KC257444, KC257445 and KC257446).
The predicted protein sequences were aligned to construct a cladogram for determining the relative position of each cloned PYVV isolate derived from the vector extracts (Fig. 4). This included the AEC22851.1 sequence from a PYVV isolated from the Colombian S.phureja, and the CP gene sequence reported for Citrus tristeza virus (CTV) (accession code NP-042867.1) was used for the base of the cladogram. Both PYVV and CTV belong to the Closteroviridae family.
The phylogenetic analysis of the clones obtained from the whitefly extracts studied (whiteflies feeding on different plants or individual symptomatic leaves) revealed slight differences (Fig. 4). These findings could support the hypothesis stating the occurrence of PYVV virus variants, not only amongst different field potato plants (11, 13) but also in a single infected plant (intra-isolates), as it was suspected in other work done by our group (31).
DISCUSSION
The present study demonstrated the PYVV molecular detection from RNA extracts of the natural Trialeurodes vaporariorum vector for the first time by using a simple method based on the amplification and sequencing of PYVV CP and Hsp70h genes.
Previous reports had confirmed the whitefly Trialeurodes vaporariorum as a natural vector of PYVV by classical plant to plant transmission protocols (2). Other viruses have been detected from their vector RNA or DNA extracts (23, 24), but the authors used approaches different from the simple one described in this study to obtain viruliferous whiteflies.
No RT-PCR bands were obtained from 1 or 2 whiteflies, possibly because it was not certain that one specific whitefly had fed on the infected leaf, whereas using pools of 3 to 20 whiteflies, it was very likely that some whiteflies had fed correctly on the infected leaf and had thus acquired the PYVV during the feeding period. No bands were obtained from either healthy plant tissue or aviruliferous whiteflies maintained on zucchini plants.
Amplicons of around 790 bp were obtained for CP gene by RT-PCR from viruliferous T. Vaporariorum extracts, close to the predicted 758 bp genome CP size reported for leaf extraction (13). Fragments of around 1,650 bp were amplified with the PYVV Hsp70h primers (Fig. 3b). Amplicons from each assay were sequenced to confirm the PYVV origin. CP gene sequence and alignments analysis had>99% similarity with those reported for PYVV isolates derived from potato symptomatic leaf-extracts. CP genes sequences obtained from the vector extract clustered in a different clade from that reported for Peruvian and Colombian CP genes accession extracted from a leaf harvested in Marinilla region (Colombian Antioquia State).
The viral RNA extracted from the vector was positive from pools of 3 to more than 20 whiteflies. Unfortunately, it was difficult to obtain positive RT-PCR from only one or 3 whiteflies, but other studies would be necessary to improve this option.
The RT-PCR amplification band was weak when RNA extracts from fewer than 5 flies were tested. The amount of viral particles detectable by RT-PCR is limited to viruses associated with the fly stylets in a semi-persistent transmission (33), contrasting with viruses transmitted in a circulatory, replicative and persistent manner (32, 34). It should be noticed that whiteflies were chilled before being removed from the leaves so that if the whiteflies «fell asleep» or became immobile due to cold while still feeding, their stylets could have been broken when they were taken from the leaves.
Whiteflies acquired the virus with its capsid; the free viral RNA titres could thus have been very low. This could explain the differences in whitefly RNA Hsp70h band intensity in contrast with the bands amplified from plant tissue-derived RNA (Fig. 3b) where the virus was replicating and was naked, meaning that the viral titres in the plant tissue extracts could have been higher than in the vector.
The amplified band from a single fly was undetectable; however, it would be important to obtain this so that information from one individual fly could be obtained by sequencing its corresponding clones. Whiteflies can transmit some viral variants but not all of them, as it was demonstrated for specific CTV variants in aphid transmission (35). Our hypothesis must thus be verified in other studies. Limitation could have been due to the low viral titre of the vector because as virions do not replicate in semi-persistent transmission, the virus does not circulate through the insect invading the salivary glands, and it can become lost in a very short period of time (32). Even more, some of the flies could have died in the feeding period. However, the simple acquisition method presented in this study can be useful for different genes characterization and for other viruses or vectors analysis, as well as for transmission and epidemiological studies.
Other viral genes could be easily studied in PYVV-harbouring viruliferous flies as it has been demonstrated for CP or Hsp70h genes. High quality viral RNA extracted from a vector will allow amplification of any gene present in the viral or vector genome. Including an internal vector control assay for amplifying the T. vaporariorum18S rRNA gene is important for confirming successful RNA extraction and avoiding false negative results.
Virus detection in RNA extracts of insect vectors has been reported for other Closteroviruses, such as CTV extracted from aphids using RT-PCR (36) and Cucurbit yellow stunting disorder virus in B. tabaci (24). Many factors contribute towards Crinivirus epidemiology, viral emergence and dominance, including host viral titre (21). Methodologies must thus be developed for studying and understanding PYVV-vector-plant interactions, as well as their epidemiology.
This is a first report of PYVV molecular detection in its natural viruliferous vector using a very simple technique for viral acquisition to obtain viruliferous vectors. This approach can be useful to be applied in future studies promoting an efficient management practice, such as those on vector acquisition and transmission, gene characterisation and host-insect vector interactions.
CONCLUSION
A molecular study was conducted for the first time to detect the presence of PYVV in RNA extracts derived from viruliferous whiteflies T.vaporariorum, the natural PYVV vector. The molecular detection involved the RT-PCR amplification of two viral genes (CP gene and Hsp70h), their sequencing and alignment analysis. The study included a simple method for obtaining viruliferous whiteflies through viral acquisition from a single symptomatic potato leaf kept in water-containing beakers. This molecular study concerning PYVV detection in its natural vector provides a useful model for studying other genes and viruses and their natural vectors as well, and represents an easy way for developing studies on viral and vector gene characterization and epidemiology.
ACKNOWLEDGEMENTS
We are grateful to Universidad Nacional de Colombia, Instituto de Biotecnología, and the Agronomy Faculty for providing the laboratory and greenhouse facilities; to Liliana Franco, Universidad Militar Nueva Granada, for providing some vector insects, and Anne-LiseHaenni, who kindly revised the manuscript and made important suggestions. A grant from Colciencias Project 2011-2013 is gratefully acknowledged.
REFERENCES
1. Ortega E, Rodríguez Y. El virus del amarilleo de las venas de papa (PYVV). INIA Divulga. 2005;4:33-36. Available in http://sian.inia.gob.ve/repositorio/revistas_tec/inia_divulga/numero%204 /ortega_e.pdf.
2. Salazar LF, Muller G, Querci M, Zapata JL, Owens RA. Potato yellow vein virus. Its host range, distribution in South America and identification as a Crinivirus transmitted by Trialeurodes vaporariorum. Ann Appl Biol. 2000;137(1):007-019.
3. Franco-Lara L, Rodríguez D, Guzmán-Barney M. Prevalence of potato yellow vein virus (PYVV) in Solanum tuberosum Group Phureja fields in three states of Colombia. Am J Potato Res. 2013;90(4):324-330.
4. Guzmán-Barney M, Franco-Lara L, Rodríguez D, Vargas L, Fierro J. Yield losses in Solanum tuberosum group Phureja cultivar Criolla Colombia in plants with symptoms of PYVV in field trials. Am J Potato Res. 2012;89(6):438-447.
5. Martelli GP, Ghanem-Sabanadzovic N, Agranovsky AA, Al Rwahnih M, Dolja VV, Dovas CI, et al. Taxonomic revision of the family Closteroviridae with special reference to the grapevine leaf roll-associated members of the genus Ampelovirus and the putative species unassigned to the family. J Plant Pathol. 2012;94(1):7-19.
6. Dolja V, Kreuze J, Valkonen J. Comparative and functional genomics of Closteroviruses. Virus Res. 2006;117(1):38-51.
7. Karasev AV. Genetic diversity and evolution of Closteroviruses. Annu Rev Phytopathol. 2000;38:293-324.
8. Livieratos IC, Eliasco E, Muller G, Olsthoorn RCL, Salazar LF, et al. Analysis of the RNA of Potato yellow veinvirus: evidence for a tripartite genome and conserved 3'-terminal structures among members of the genus Crinivirus. J Gen Virol. 2004;85(7):2065-2075.
9. Eliasco E, Livieratos IC, Muller G, Guzmán M, Salazar LF, Coutts RHA. Sequences of defective RNAs associated with Potato yellow vein virus. Arch Virol. 2006;151(1):201-204.
10.Guzmán M, Rodríguez P. Susceptibility of Solanum phureja (Juz. EtBuk) to Potato yellow vein virus. Agron Col. 2010;28(2):219-224.
11.Guzmán M, Ruiz E, Arciniegas N, Coutts RHA. Occurrence and variability of Potato Yellow Vein Virus in three departments of Colombia. J Phytopathol. 2006;154(11-12):748-750.
12.López R, Acensio C, Guzmán M, Boonham N. Development of real-time and conventional RT-PCR assays for detection of Potato yellow vein virus. J Virol Methods. 2006;136(1-2):24-29.
13.Offei SK, Arciniegas N, Muller G, Guzmán M, Salazar LF, Coutts RHA. Molecular variation of Potato yellow vein virus isolates. Arch Virol. 2004;149(4):821-827.
14.Guzmán-Barney M, Hernández AK, Franco-Lara L. Tracking Foliar Symptoms Caused by Tuber-Borne Potato Yellow Vein Virus (PYVV) in Solanum phureja (Juz et Buk) Cultivar «Criolla Colombia». Am J Potato Res. 2013;90(3):284-293.
15.Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR, Daszak P. Emerging infectious diseases of plants: Pathogen pollution, climate change, and agrotechnology drivers. Trends Ecol Evol. 2004;19(10):535-544.
16.Tzanetakis IE, Martin R, Wintermantel WM. Epidemiology of criniviruses: an emerging problem in world agriculture. Front Microbiol. 2013;4:119-124.
17.Jones DR. Plant viruses transmitted by whiteflies. Eur J Plant Pathol. 2003;109(3):195-219.
18.Wintermantel WM. Emergence of greenhouse whitefly (Trialeurodes vaporariorum) transmitted criniviruses as threats to vegetable and fruit production in North America. 2004. Online. Available in http://www.apsnet.org/publications/apsnetfeatures/Pages/GreenhouseWhitefly.aspx. Accessed Jul, 2013.
19.Brown JK, Czosnek H. Whitefly transmission of plant viruses. In: Plumb RT (Ed). Advances in Botanical Research. Academic Press, San Diego, CA, USA. 2002; pp 65-100.
20.Gottlieb Y, Zchori-Fein E, Mozes-Daube N, Kontsedalov S, Skaljac M,Brumin M, et al. The transmission efficiency of Tomato Yellow Leaf Curl Virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. J Virol. 2010;84(18):9310-9317.
21.Wintermantel WM, Cortez A, Anchieta A, Gulati-Sakhuja A, Hladky L. Co-infection by two Criniviruses alters accumulation of each virus in a host-specific manner and influences efficiency of virus transmission. Phytopathology. 2008;98(12):1340-1345.
22.Wintermantel WM, Wisler GC. Vector specificity, host range, and genetic diversity of Tomato Chlorosis Virus. Plant Dis. 2006;90(6):814-819.
23.Gil-Salas FM, Morris J, Colyer A, Budge G, Boonham N, Cuadrado IM, et al. Development of real-time RT-PCR assays for the detection of Cucumber vein yellowing virus (CVYV) and Cucurbit yellow stunting disorder virus (CYSDV) in the whitefly vector Bemisia tabaci. J Virol Methods. 2007;146(1-2):45-51.
24.Ruiz L, Janssen D, Velasco L, Segundo E, Cuadrado IM. Quantitation of cucurbit yellow stunting disorder virus in Bemisia tabaci (Genn.) using digoxigenin-labelled hybridisation probes. J Virol Methods. 2002;101(1-2):95-103.
25.Gamarra H, Chuquillanqui C, Müller G. Transmisión del virus del amarillamiento de las venas de la papa en variedades y clones de Solanum tuberosum L. Potato international center (CIP); Entomological National Convention, Lima, Perú. 2002.
26.Cubillos K. Determination of virus variants of potato yellowing vein virus (PYVV) by molecular analysis of three genes in colombian isolates of Solanum spp. Master degree Thesis. Sciences Faculty. Universidad Nacional de Colombia. 2011. Available in http://www.bdigital.unal.edu.co/4193.
27.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792-1797.
28.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465-9.
29.Castresana J. Selection of conserved blocks from multiple alignmentsfor their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540-552.
30.Wang HC, Li K, Susko E, Roger AJ. A class frequency mixture model that adjusts for site-specific amino acid frequencies and improves inference of protein phylogeny. BMC Evol Biol. 2008;8(1):331.
31.Chaves-Bedoya G, Guzmán-Barney M, Ortiz-Rojas L. Genetic heterogeneity and Evidence Of Putative Darwinian Diversifying Selection In Potato Yellow Vein Virus (PYVV). Rev Agron Col. 2013;31(2):161-168.
32.Chen A, Walker GP, Carter D, Ng JCK. A virus capsid component mediates virion retention and transmission by its insect vector. PNAS. 2011;108(40):16777-16782.
33.Fereres A, Moreno A. Behavioral aspects influencing plant virus transmission by homopteran insects. Virus Res. 2009;141(2):158-168.
34.Andret-Link P, Fuchs M. Transmission specificity of plant viruses by vectors. J Plant Pathol. 2005;87(3):153-165.
35.Velazquez-Monreal JJ, Mathews DM, Dodds JA. Segregation of distinct variants from Citrus tristeza virus Isolate SY568 using aphid transmission. Phytopathology. 2009;99(10):1168-1176.
36.Saponari M, Manjunath K, Yokomi R. Quantitative detection ofCitrus tristeza virus in citrus and aphids by real-time reverse transcription-PCR (TaqMan). J Virol Methods. 2008;147(1):43-53.
Recibido: 31-1-2014.
Aceptado: 11-8-2014.
*Correspondent: Mónica Guzmán-Barney. E-mail: mmguzmanb@unal.edu.co.