Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista de Protección Vegetal
versión impresa ISSN 1010-2752versión On-line ISSN 2224-4697
Rev. Protección Veg. vol.30 no.2 La Habana mayo.-ago. 2015
SHORT COMMUNICATION
Pyramiding TYLCV and TSWV resistance genes in tomato genotypes
Pirimidación de genes de resistencia a TYLCV y TSWV en genotipos de tomate
Olimpia Gómez ConsuegraI, Mayte Piñón GómezI, Yamila Martínez ZubiaurII
IHorticultural Research Institute Liliana Dimitrova(IIHLD), Carretera Quivicán Km 33½, Mayabeque, Cuba.
IINational Center for Animal and Plant Health (CENSA), Apartado 10. San José de las Lajas, Mayabeque, Cuba. E-mail: yamila@censa.edu.cu.
ABSTRACT
The Cuban tomato line LD3 , in which the Ty-1 gene from Solanum chilense LA 1969 was detected, was crossed with an F1 hybrid carrying the genes Ty-3 from S. chilense LA 2779 and Sw-5 from S. peruvianum. A hundred F2 plants were transplanted to a ferralitic soil in the field and the F2 plants individually evaluated in the field with high natural TYLCV-IL (Cu) incidence by using a scale from 0 (no symptom) to 4 (severe stunting). The total DNA extracted from the F2 plants were analyzed per individual plant by using PCR markers linked to the genes Ty-3 (resistance to TYLCV) and Sw-5 (resistance to TSWV) so that both genes could be detected in a same genotype. Eighty two F2 plants showed no symptoms under natural TYLCV incidence (0 symptoms) with a productivity ranging from 1.6 to 9.7 kilograms per plant. The Principal Component Analysis (PCA) permitted grouping the different genotypes. Six F2 plants showed the Ty-3 gene in homozygosis and 12 in heterozygosis; 33 F2 plants showed the Sw-5 gene according to their DNA analysis. Three F2 plants (number 65, 68 and 78) showed the Ty-3 and Sw-5 genes in homozygosis besides the Ty-1 gene (resistance to TYLCV) from their female parent. Their productivity ranged from 6.0 to 6.8 kilograms per plant. These are the first Cuban results in which resistance genes for two important diseases in the tomato crop are obtained in a same genotype by the early and simultaneous molecular screening for resistance and the productivity evaluation under field conditions, Both tools, when simultaneously used, are efficient in reducing selection time and space.
Key words: TYLCV, TSWV, pyrimiding, resistance genes, tomato, genotypes.
RESUMEN
En este estudio, los cruzamientos se realizaron entre la línea cubana LD3, con gen de resistencia Ty-1, procedente de Solanum chilense LA 1969, y el híbrido F1 portador de los genes Ty-3 de S. chilense LA 2779 y Sw-5 de S. peruvianum. Se obtuvieron 100 plantas F2 que fueron trasplantadas a campo, en suelo ferralítico rojo. Las plantas F2 fueron evaluadas en condiciones naturales de campo en áreas con alta presión de TYLCV-IL (Cu). Las plantas individuales fueron evaluadas con escala de síntomas desde 0 (sin síntomas) a 4 (enanismo severo). Los ADN totales extraídos fueron analizados en plantas individuales de la población F2 en estudio, utilizando marcadores PCR ligados a los genes Ty-3 (resistencia a TYLCV) y Sw-5 (resistencia a TSWV) con el objetivo de detectar estos genes en un mismo genotipo. Un total de 82 plantas F2 no mostraron síntomas bajo condiciones naturales de incidencia de TYLCV-IL (Cu) (grado 0) con un rango de productividad de 1.6 a 9.7 kilogramos por plantas. El Análisis de Componentes Principales permitió agrupar los genotipos. En seis plantas se detectó la presencia de gen Ty-3 en homocigosis y en 12 plantas en heterocigosis; 33 plantas F2 demostraron el gen Sw-5 a partir de los análisis de ADN realizado. En tres plantas F2 (números 65, 68 y 78) se encontraron en homocigosis los genes Ty-3 y Sw-5 en adición del gen Ty-1 (resistencia a TYLCV) procedente del parental femenino. El rango de productividad obtenido fue de 6.0 a 6.8 kilogramos por plantas. Estos son los primeros resultados en Cuba donde se obtienen genes de resistencia en un mismo genotipo para dos importantes enfermedades del cultivo del tomate con una evaluación simultánea y temprana de la resistencia y la productividad en condiciones de campo, mediante el uso temprano de marcadores moleculares en la reducción del tiempo y el espacio en el proceso de selección de genotipos promisorios.
Palabras clave: TYLCV, TSWV, pirimidación, genes de resistencia, genotipos, tomate.
Begomoviruses, mainly the Tomato yellow leaf curl virus (TYLCV), and tospoviruses, such as Tomato spotted wilt virus (TSWV), Tomato Chlorotic spot virus (TCSV) and Groundnut ring spot virus (GRSV), are major diseases in the tomato (Solanum lycopersicum L) crop. In Cuba, TYLCV became the most important tomato disease since the last eighties when it appeared in the country (1). On the contrary, tospovirus species has not been reported in the Island, but it is a major constraint in different neighboring countries.
Different TYLCV resistance genes have been reported to provide a broad spectrum of protection to many begomovirus species that are wide spread in the world (2). Zamir et al. (3) were the first to report a major incompletely dominant gene (Ty-1) on chromosome 6 derived from Solanum chilense (Dunal) Reiche LA 1969. Plants homozygous for S. chilense alleles at TG297 and TG97 markers on chromosome 6 were symptomless, while susceptible control had severe symptoms. The introgression of this gene provides high resistance to a broader array of begomoviruses to some Cuban parent lines (4). Ji and Scott (5) reported a partially dominant gene also on the long arm of chromosome 6, further designated Ty-3 gene (6, 7) in lines derived from S. chilense LA2779. The gene Sw-5 confers resistance to TSWV in a dominant way (8)
A breeding program was needed at the Horticultural Research Institute «Liliana Dimitrova» (IIHLD) supported by the National Center for Animal and Plant Health (CENSA), to improve resistance level by pyramiding genes for TYLCV and TSWV resistance in high yielding genotypes to enhance a sustainable production. The aim of the present work was to incorporate different TYLCV resistance genes (Ty-1 and Ty-3) and the Sw-5 gene (resistance to TSWV) into a single tomato genotype to be screened by molecular markers in early generations and to establish the relationships between TYLCV resistance and productivity traits.
The cross of the genetic material was made between LD3, a Cuban tomato line in which the Ty-1 gene from S. chilense LA 1969 was detected by Piñón et al. (9), and an F1 hybrid carrying the Ty-3 gene from S. chilense LA 2779 and the Sw-5 gene from S. peruvianum. In a field trial, 100 F2 plants were transplanted to ferralitic soil at the IIHLD. The distance between the plants was 0.25 m with an interrow space of 1.40 m, respectively. No chemical treatment was applied to control Bemisia tabaci Genn., the TYLCV-vector.
After transplanted, the F2 plants were evaluated under high natural TYLCV incidence in open field conditions with no escape to the susceptible control cv. `Campbell 28'. The plants were individually evaluated weekly until 56 days after inoculation by using a scale from 0 (no symptom) to 4 (stunting) (10).
The plant DNA was extracted using a short nucleic acid extraction technique (11). Leaf tissue (0.15 g) was macerated in liquid nitrogen followed by the addition of the extraction buffer (100 mM Tris-HCL pH8, 50 ml NaCl, 1% SDS, 10 ml ß-mercaptoethanol). The final product was suspended in 300 ml of TE buffer (10 mÌ Tris-HCl, pH8 1 ml EDTA). The total DNA extracted from the F2 plants were analyzed per individual plant by using PCR markers linked to the genes Ty-3 (7) and Sw-5 (8) so that both genes could be detected in a same genotype. The individual plants carrying these genes were quantified. The PCR-fragments were visualized with UV light and purified from PCR reaction according to the standard protocol (Wizard Plus SV miniprep DNA purification System). The sequencing was conducted under BigDyeTM terminator cycling conditions (Macrogen Service).The Correlation and Principal Component analyses were carried out using SAS 8.1 software (SAS Institute, Cary, N. C. USA), in which variables such as TYLCV severity, number of flowers, fruits and fruit set at the first four bunches, number of bunches and number of total fruits per plant; mean fruit weight and yield per plant were included.
Differences for TYLCV severity and for productivity characters were detected among genotypes. Eighty two F2 plants showed no symptoms under natural TYLCV incidence (0 symptoms) with a productivity ranging from 1.6 to 9.7 kilograms per plant making possible to select for TYLCV resistance and high productivity. (Table 1)
TYLCV severity was negatively and significantly correlated with the flower number at the first four bunches in the plant (r = -0.783). It also decreased other productivity traits such as: fruit and bunch numbers per plant (r = -0.886 and -0.8334, respectively), fruit set (r = -0.93) and yield per plant (r = -0.893), which explained the productivity decrease observed in this work when TYLCV severity increased (table 1). Gómez et al. (9) informed these effects in comparing resistant and susceptible tomato cultivars in tropical conditions. Mejias et al. (12) found that TYLCV infection led to yield loss mainly due to the reduction of the number of fruits per plant.
According to the Principal Component Analysis (PCA), where the first three components extracted 60% of the total variability, the genotypes could be grouped. The F2 plants numbers 65, 68 and 78 were included into the 1st group highlighting for symptomless and high yield (6.0 to 6.8 kilogram per plant).
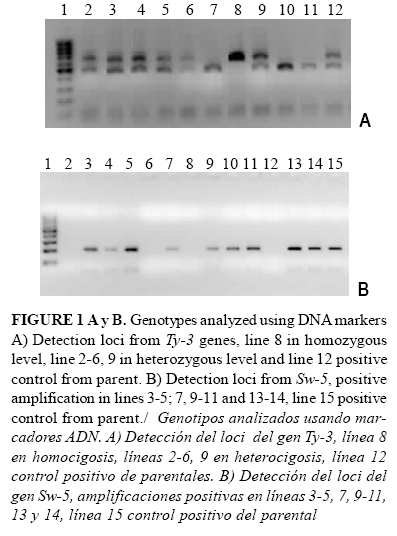
Six F2 plants showed the Ty-3 gene in homozygosis with an amplified fragment of aprox. 640pb; twenty in heterozygosis and an amplified fragment of aprox. 500 and 640pb (Figure 1A), and 33 F2 plants showed the Sw-5 gene according to DNA analyses (Figure 1B).
The sequence in forward and reverse direction of the 629 pb from the fragment showed 100% of identity with Ty-3 gene from Gc43 Genotype resistant to begomoviruses in Guatemala (12).
The transfer of the gene Sw-5 was observed for the amplification of PCR-fragments about 200pb with 100% of identity to Sw-5 a gene from Genebank (AY007366.1).
Three F2 plants (number 65, 68 and 78), symptomless and with high productivity, showed the Ty-3 and Sw-5 genes in homozygosis besides the Ty-1 gene (resistance to TYLCV) previously reported by Piñón et al. (2) in their female parent.
DNA marker technology has been used in commercial plant breeding programs since the early 1990's and has proved helpful for rapid and efficient transfer of useful traits into agronomically desirable varieties and hybrids. In this work the main goal was achieved through the early molecular screening for resistance and the productivity evaluation under field conditions; both tools, when simultaneously used, showed to be efficient in reducing selection time and space.
REFRENCES
1. Martínez Y, Martínez MA, Quiñones M, Miranda I, Holt J, Chancellor T. Estudio de factores que influyen en la epifitología del complejo mosca blanca-geminivirus, en la región oriental de Cuba. Rev Protección Veg. 2009;24(1):47-50.
2. Ji Y, Scott JW, Hanson P, Graham E, Maxwell DP. Sources of resistance, inheritance, and location of genetic loci conferring resistance to members of the tomato-infecting begomoviruses Chapter 2. In H. Czosnek (ed.), Tomato Yellow Leaf Curl Virus Disease. 2007. pages 343-362.
3. Zamir D, Michelson I, Zakay Y, Navot N, Zeidan M, Sarfatti M, et al. Mapping and introgression of a Tomato yellow leaf curl virus tolerance gene, Ty-1. Theor Appl Genet. 1994;88:141-146.
4. Piñón M, Gómez O, Cornide MT. RFLP analysis of Cuban tomato breeding lines with resistance to Tomato yellow leaf curl virus. Acta Horticulturae. 2005;695:273-276.
5. Ji Y, Scott JW. Identification of RAPD markers linked to Lycopersicon chilense derived begomovirus resistant genes on chromosome 6 of tomato. Acta Horticulturae. 2005;695:407-416.
6. Ji Y, Scott JW. Development of SCAR and CAPS markers linked to tomato begomovirus resistance genes introgressed from Lycopersicon chilense. HortSci. 2005;40:1090.
7. Ji Y, Schuster DJ, Scott JW. Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Molecular Breeding. 2007;20(3):271.
8. Langella R, Ercolano MR, Monti L, Frusciante L, Barone A. Molecular marker assistance transfer of resistance to TSWV in tomato elite. J Hort Sci & Biotech. 2004;79(5):806-810.
9. Gómez O, Piñón M, Martínez Y, Quiñones M, Fonseca D, Laterrot H. Breeding for resistance to begomovirus in tropic-adapted tomato genotypes. Plant Breeding. 2004;123:275-279.
10.Scott JW, Schuster DJ. Screening of accessions for resistance to the Florida tomato geminivirus. 1991; TGC Report 41.
11.Noris EE, Hidalgo GP, Moriones E. High similarity among the tomato yellow leaf curl isolates from the west Mediterranean Basin: the nucleotide sequence of an infectious clone from Spain. Arch Virol. 1994;135:165-170.
12.Mejía L, Teni RE, Vidavski F, Czosnek H, Lapidot M, et al. Evaluation of tomato germplasm and selection of breeding lines for resistance to begomoviruses in Guatemala. Acta Horticulturae. 2005;695:251-255.
Recibido: 23-12-2014.
Aceptado: 13-2-2015.