My SciELO
Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Biotecnología Aplicada
On-line version ISSN 1027-2852
Biotecnol Apl vol.26 no.1 La Habana Jan.-Mar. 2009
REVIEW
Challenges for the design of effective vaccines or drugs against Cholera and Tuberculosis: Possible contributions from omics and bioinformatics
Contribuciones de la bioinformática en el diseño de nuevas vacunas y fármacos contra el cólera y la tuberculosis.
Yuliet Mazola, Arielis Rodríguez, Rolando Rodríguez
Chemistry-Physics Division, Center for Genetic Engineering and Biotechnology, CIGB. Ave 31 / 158 and 190, Cubanacan, Playa, PO Box 6162, Havana, Cuba
ABSTRACT
This manuscript shows the current situation of two re-emerging diseases: cholera and tuberculosis, which still constitute one of the major health problems in poor countries. In this regard, difficulties, challenges and perspectives for the development of new drugs and vaccines against these diseases are discussed. The possible contributions of structural biology, bioinformatics and omics to obtain an effective treatment are also considered.
Keywords: Tuberculosis, cholera, drug design, bioinformatics, vaccine, pathogenesis, proteomics, genomics.
RESUMEN
El cólera y la tuberculosis son enfermedades re-emergentes que constituyen un serio problema de salud en los países pobres. En este trabajo se realiza una revisión sobre las dificultades, los retos y las perspectivas para el tratamiento de ambas enfermedades. Particularmente se abordan las aplicaciones de la biología estructural, la bioinformática y la integración de datos en el desarrollo de nuevas drogas y vacunas.
Palabras clave: tuberculosis, cólera, diseño de drogas, bioinformática, vacunas, patogénesis, proteómica, genómica.
INTRODUCTION
Cholera and tuberculosis (TB) are infectious diseases declared as global emergencies by the World Health Organization (WHO) (1, 2). Emerging disease are those infections that either are newly appearing in the population or are rapidly increasing in incidence or expanding in geographical range (3). The etiological agents responsible for cholera and tuberculosis were identified during 19th century; they are Vibrio cholerae and Mycobacterium tuberculosis respectively. Thus, these are not precisely new diseases, although new variants of both microorganisms have emerged like the serogroup O139 of V. cholerae and the multidrug resistant (MDR) strains of M. tuberculosis. Instead, they are infectious diseases that still remain with high incidence and lethality in a significant part of the world, particularly in underdeveloped countries.
In 2006, a total of 52 countries officially reported WHO 236 896 cases of cholera, including 6311 deaths (4). However, the incidence of the disease is much higher since a considerable number of cases are not reported (4). Even in this scenario, in 2006 the case fatality rate increased from 1.72% estimated in 2005 to 2.66% (4). On the other hand, one-third of the worlds population is currently infected with the TB bacillus. Even though only 5-10% of the individuals infected with M. tuberculosis (but no with the human immunodeficiency virus, HIV) become sick, TB is a major cause of illness and death worldwide. In fact, only in 2006 WHO reported 9.2 million new cases and 1.7 million deaths, of which 0.7 million cases and 0.2 million deaths corresponded to HIV-positive people (5). Unfortunately, despite all the efforts that have been made, no effective treatment is available to completely prevent and cure the infection by both pathogens.
This article points out how different applications of bioinformatics may contribute to the development of new treatments for these diseases. In the case of cholera, bioinformatics could be used to improve the understanding about environmental survival and pathogenesis of V. cholerae. Therefore, data integration of different omics results and sequence analysis may accelerate the discovery of new vaccines and contribute to find new strategies to combat the disease. For TB, a different approach on bioinformatics is presented. It focuses on the rational design of novel and better drugs based on available 3D structures, either of the target or already known anti-tuberculosis drugs. The necessity for developing new drugs obeys mostly to the emergence of strains that are resistant to traditional chemotherapy. In this sense, computer-based drug design may allow the identification of new drugs with different mechanisms of action to overcome resistance.
CHOLERA
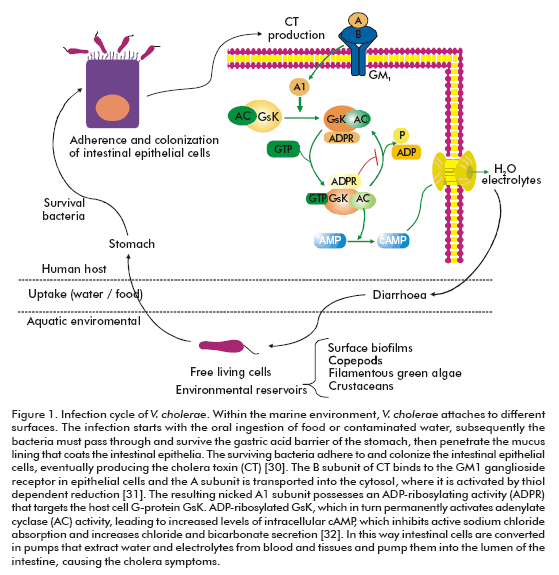
Cholera is characterized by a profuse watery diarrhea caused by the cholera toxin of V. cholerae (Figure 1). The dehydration is of such magnitude that, without treatment, causes hypotensive shock and leads to death within hours (6). Although more than 200 serogroups of V. cholerae have been described, cholera is associated only with the O1 and O139 serogroups (6). The O1 serogroup is divided into three serotypes: Ogawa, Inaba and Hikojima, which are further divided into two biotypes, classical and El Tor. Since the nineteenth century, seven pandemics of cholera have occurred (7, 8).
Treatment and prevention
Clinical management of cholera has considerably advanced; in fact, the case fatality rate decreased with the development of the rehydration therapy (9). In addition, antimicrobial agents are used to reduce the duration of the symptoms (10). However, the number of drugs that can be used in the treatment has decreased due to the emergence of V. cholerae strains resistant to multiple antibiotics (11, 12). Cholera is a disease of rapid onset and propagation; therefore, public health systems are easily overwhelmed when a cholera outbreak occurs, especially in the poorest countries (13). Several control measures like improvements in water and sanitation are recommended for cholera prevention, but these are real challenges for developing countries. Thus, a successful prevention of cholera can only be achieved through vaccination (14). Undoubtedly, this field has shown increasing results, but today an effective vaccine with a broad spectrum of action, few side effects and an economic production is not available yet.
The first licensed vaccine against cholera was made with killed whole-cells of the V. cholerae O1 serogroup and the recombinant B-subunit of cholera toxin (WC/rBS). Although such vaccine is safe and effective, generating an immune response requires of two oral doses with an interval of 14-42 days (15, 16). Consequently, this vaccine is very expensive and has logistic difficulties, like reaching twice the same population. Besides, a large volume of liquid (150 mL) is needed for its administration, and cannot be given to children under two years of age (17).
A variant of the WC/rBS vaccine without recombinant B-subunit has been licensed in Viet Nam. This vaccine is safe and immunogenic, does not need buffer solution and may be used in young children (18). But it is also given in two doses, though cheaper than the internationally licensed WC/rBS vaccine (19). In both cases, killed O139 whole-cells were added to create bivalent killed whole-cell vaccines, which are also safe and immunogenic (20, 21).
Recent efforts to develop a vaccine against cholera have focused on the use of live attenuated strains of V. cholerae. Such strains possess the pathogenicity factors required for the colonization of the small intestine (e.g., motility, fimbriae, neuraminidase, etc) and express the B-subunit of the cholera toxin. In this way, the second internationally licensed cholera vaccine was a live attenuated genetically modified V. cholerae O1 strain of the classical biotype, named CVD 103-HgR. It is administered in a single oral dose to individuals older than two years (17). Although it is safe and has relatively low production costs (22), it did not show a convincing protection in a population exposed to cholera a long time after immunization, in a large field trial performed in Indonesia (23). Thus, even though this vaccine is licensed, it is currently unavailable because the production was stopped (17). Other oral live vaccines, against the biotype El Tor and the O139 serogroup, are under development and have been shown to be effective in preliminary studies and clinical assays (24-27).
Other alternatives to combat cholera might be designed. In this sense, a different generation of vaccines could be based on the identification of bacterial targets that stimulate the human immune response (28). Other strategies could be directed to the inhibition of cholera toxin binding to receptors in the intestine (29), the improvement in treatment options for managing the symptoms (29) or the eradication of pathogenic V. cholerae from community water supplies. An important challenge for these goals is the fact that, despite all the efforts and the achieved results, the pathogenesis of V. cholerae is not completely understood.
OVERVIEW OF V. cholera AND ITS ENVIRONMENT
The viable but not cultivable status and biofilm formation are two survival strategies of pathogenic vibrios in the marine environment. V. cholerae switches to a viable but not cultivable state in response to nutrient deprivation (33), but the exact environmental conditions serving to resuscitate these cells back to freeliving virulent organisms are unknown. The appropriate signals could be related with the transition from the aquatic system into the human intestinal environment upon ingestion (8).
On the other hand, biofilm formation depends on the synthesis of an exopolysaccharide (EPS) encoded by the Vibrio polysaccharide (vps) genes (34). The O1 El Tor strain requires, besides, the MSHA type IV pilus and flagellar motility (35, 36). Three distinct signaling pathways modulate EPS production in V. cholerae: i) a quorum-sensing pathway, wherein the absence of the transcriptional regulator HapR results in enhanced EPS synthesis and biofilm formation (37); ii) a flagellum-dependent pathway, wherein a sodiumdriven flagellar motor sense reduction in flagellar rotation and induce EPS synthesis and biofilm formation via the VpsR/VpsT signaling cascade (38); and iii) a phase variation pathway that yields two distinct morphological variants termed smoothly and rugose (wrinkled). Rugose variants are associated with an enhanced capacity to produce EPS (39). Maintaining multiple signaling pathways for EPS synthesis and biofilm formation may contribute to persistence in the dynamic aquatic environments occurring between seasonal cholera outbreaks. Although the VpsR/VpsT signaling cascade is responsible for biofilm formation, a direct interaction between the transcriptional activator, VspR or VspT, and the vps structural genes has not been demonstrated (38, 40). Curiously, both VspR and VspT are homologous to response regulators of twocomponent regulatory systems, which are typically associated with sensory histidine kinases, but yet no cognate sensor kinases have been identified (38, 40).
OVERVIEW OF V. cholerae VIRULENCE
A key virulence factor of V. cholera is the cholera toxin (CTX) genetic element containing the CT operon (ctxAB) which encodes cholera toxin (41). Another important cluster of virulence genes corresponds to the TCP pathogenicity island, also known as Vibrio pathogenicity island (42). Several gene clusters are found in the Vibrio pathogenicity island, such as TCP (toxin co-regulated pilus), ACF (accessory colonization factor) and a cluster containing three genes between tagA to tagD. The transcriptional activator ToxT is also located in this chromosome region (42) . The TCP is a type IV pilus composed of a single subunit: TcpA. This virulence factor serves as the receptor of the CTXF bacteriophage and is also required for intestinal colonization (43). TCP seems to facilitate microcolony formation on the epithelial cell surface (44). The function of some proteins coded by the Vibrio pathogenicity island, for example the gene products of tcpI and acfB, are still unknown.
The transcriptional regulation of the CTX and TCP clusters are under the control of ToxT (45). At the same time, the transcription of the toxT gene is controlled by the two transcriptional activators, ToxR and TcpP (46, 47). This regulatory cascade is known as ToxR virulence regulon. Within the human intestine (unlike in the laboratory) the ctx transcription requires toxR but not tcpP (48), and in addition de-pends on the bacteria motility and chemotaxis (49). This suggests that the true V. cholerae virulence cascade may differ significantly from that elucidated through in vitro laboratory experiments, and thus demonstrates the importance of studying pathogenesis in the context of a living host rather than in laboratory conditions.
Although additional putative virulence factors, such as neuroaminidases, secreted proteases, mannosefucose- resistant cell-associated hemagglutinin and mannose-sensitive hemagglutinin have been characterized, their exact role in the pathogenesis of V. cholerae is uncertain.
STUDIES IN OMICS AND POSSIBLE APPLICATIONS OF BIOINFORMATICS
Genomics, transcriptomics and proteomics studies have been conducted to provide a better understanding about V. cholerae. Currently, different Vibrios species genomes have been completely sequenced (Table 1). In particular, the V. cholerae genome consists of two circular chromosomes: I or large with 2.96 Mb and II or small with 1.07 Mb, which encode 2775 and 1115 ORF, respectively (50). Chromosome I encodes most of the genes essential for cell function (e.g., DNA replication, transcription, translation, etc.) and pathogenicity (e.g., toxin, surface antigens, and adhesion), whereas chromosome II encodes a larger proportion of hypothetical genes. In other Vibrio species, the vast majority of hypothetical genes are also located in the small chromosome.
Certainly, the genome sequence of V. cholerae is one of the most promising results to develop better vaccines, new diagnostics methods and treatments of cholera. Identifying the function of hypothetical proteins and the role of the small chromosome in the biology of this pathogen may be important to achieve this goal. In this sense, bioinformatics methods for sequence comparison and phylogenetic analysis are useful to suggest protein functions and study the evolution of different Vibrio species. Besides, by using comparative genomics the molecular basis of the distinct clinical behavior of different pathogenic species can be disclosed. For example, genes for the type III secretion system (TTSS) were identified in the genome of V. parahaemolyticus, but V. cholerae does not have such genes (51). Type III secretion system is a central virulence factor in bacteria that causes gastroenteritis by invading or interacting with intestinal epithe lial cells. This finding explains the different clinical features of V. parahaemolyticus and V. cholerae infections, which include inflammatory and noninflammatory diarrhea, respectively. Also by comparative genome analyses of pathogenic and nonpathogenic vibrios, for example V. fischeri, it is possible to identify features that are common to beneficial and pathogenic bacteria.
Gene expression profiles at the transcriptional level have been used to analyze V. cholerae cell growth in vivo. In a first study, Xu et al. used the rabbit ileal loop model of V. cholerae infection to obtain in vivo grown cells under exponential phase, and compared the global transcriptional pattern of these cells with those others grown under laboratory conditions (52). They suggested that iron limitation, nutrient deprivation and anaerobiosis are prominent stressing conditions experienced by the bacteria in the rabbit upper intestine. Also, intestinal environment signifi-cantly enhanced the expression of several virulence genes, including those involved in processes like mo-tility, chemotaxis, intestinal colonization and toxin production. In a second study, V. cholerae cells shed from cholera patients were compared with V. cholerae cells grown in LB to stationary phase (56). In this case, a high expression level of those genes required for nutrient acquisition and motility was also found, but the genes required for chemotaxis were under repression and, besides, no differential expression was found for genes of the ToxRS/TcpPH/ToxT regulon. The authors of this study suggested that human colonization creates a hyper-infectious bacterial state that is maintained after dissemination and that may contribute to epidemic spread of cholera.
Moreover, proteomics has been used to understand the physiological adaptation of V. cholerae to different conditions. In particular, the effect of mild acid treatment on the physiology of the classical strain O395 was investigated (57). Also the proteome of El Tor strain N16961 was compared in aerobic and anaerobic conditions in the pI range of 2 to 11, the anaerobic condition was used as an approximation to the environment found by V. cholerae during infection in the intestine (58). Most of the proteins analyzed in this case belong to the aerobic or anaerobic metabolism of carbohydrates (58). On a recent report, a more realistic approximation to host conditions was achieved by analyzing the proteome of V. cholerae recovered from human stool (59). In this case, the majority of the identified proteins are involved in protein synthesis and energy metabolism (59), in correspondence with previous studies. Proteins related with the pathogenesis of cholera, including the A and B subunits of CT and the toxin-coregulated pilus, were also identified. The outer membrane porin, OmpU, was identified in all the analyzed samples (59). Furthermore, a proteome reference map was presented for El Tor strains in the pI range of 4 to 7 (60).
Now it is widely accepted that a catalogue of genes, transcripts or proteins is not enough to understand and the biological system (61). To improve our knowledge about V. cholerae the available data must be integrated. Bioinformatics can support such integration process with database searching, text mining tools, network construction and pathway analysis. At the same time further investigations are needed, especially in rela-tion with the environmental persistence mode and the virulence process of V. cholerae.
TUBERCULOSIS
TB infection is mainly asymptomatic. It is generally controlled by the immune system responses but a residual population of latent mycobacteria may persist. About 5-10% of latent infection may progress to active disease (5). The leading reasons are the weakness of immune systems responses, frequently in people infected with HIV, and the inefficient control of the initial infection or a subsequent reinfection. The active disease or post-primary TB is predominantly a pulmonary disease involving extensive damage to the lungs and efficient aerosol transmission of bacteria.
Treatment and prevention
TB might be cured with chemotherapy if it is early detected and fully treated; instead the death rate is about 50% (62). Two disadvantages of this treatment are its time length and its ineffective elimination of persistent bacilli (63). Certainly, patients may delay in medical attention which inhibits its fast detection and treatment, increasing the chance of transmission, complications and death. Besides, poor people which are the most vulnerable population have limited access to therapy. Latent infection is treated using a single antibiotic (usually isoniazid) during 6-9 months. In contrast, the active disease has been treated with combination of several antibiotics for over fifty years to avoid the rapid development of resistance (64). First-line drugs include isoniazid, rifampin, pyrazinamide and ethambutol, given during six months. Secondline drugs such as para amino-salicylate, kanamycin, fluoroquinolones, capreomycin, ethionamide and cycloserine are used when therapy fails because of bacterial drug resistance or intolerance to one or more drugs (65). TB prevention and control involves the identification and treatment of infected persons, including their contacts, and vaccination. The current vaccine against TB, known as Bacille Calmette Guerin (BCG) is derived from Mycobacterium bovis which is a very close relative of M. tuberculosis. BCG vaccines protective efficacy is still a matter of debate (66). It is effective in preventing childhood TB, including the meningeal and miliary or disseminated forms of the disease, but it fails to protect against the predominant pulmonary form of the disease in adults (67-69), which is the most prevalent one. Sacksteder and Nacy pointed out that BCG has undergone significant genetic changes since its development in the early twentieth century and these variations in relation to M. tuberculosis genome could play a significant role in the inadequate efficacy of BCG vaccination in the protection against pulmonary TB (70). Two other drawbacks of BCG vaccine have limited its use. First, it interferes with the interpretation of the purified protein derivative test, the only skin test available for rapid TB diagnosis. Secondly, it is not recommended for boosting vaccination because it is a live attenuated virus vaccine (71). As a result, some countries in the world, like the US and the Netherlands, do not vaccinate with BCG (70). These countries prefer to use active case detection, treatment and surveillance with the purified protein derivative test, to control and monitor M. tuberculosis exposure in their populations. In Europe and Japan, BCG vaccination is a voluntary choice for parents (70).
For many years, the scientific community thought that TB was controlled using BCG and therefore it would be eradicated soon. Because of that, very few new TB vaccines have been developed and tested in humans (70). But, such way of thinking was completely wrong. Nowadays, three major problems are directly implied in the rise of TB incidence, deaths and therapeutic failure. The first is drug resistance mainly caused by a prolonged therapy, and also the use and often misuse of antibiotics, leading to the evolution of MDR strains of M. tuberculosis (72).
WHO reported an estimated 0.5 million deaths associated with MDR strains infection (5). The second problem is related to the spread of HIV infection (73), which depresses and deteriorates the immune system. The last but not least, is the abandonment of TB control programs.
At present, many researchers are hopingly testing different forms of recombinant BCG waiting for modifications to improve its protective effects (74, 75). The first human clinical trial of a recombinant BCG vaccine (rBCG30) was initiated in 2004 (76). This is a live vaccine, consisting of BCG genetically modified to produce abundant amounts of a 30 kDa antigen that has been shown to produce a strong immune response in animals and humans. Other live organisms have been considered as potential TB vaccines; one of these is Mycobacterium microti. This candidate vaccine orally administered in mice demonstrated a greater efficacy as compared to BCG at a high dose (77).
The experts' opinion suggests that the entire characteristic required to develop an effective vaccine against TB may not be found in a single vaccine, instead multiple vaccines may be needed for an efficient control of the epidemics. They recommended a hybrid approach using multiple vaccines that can be administered regardless of the infection status of the individual and with activity both in naive and already infected individuals (70, 78).
Certainly, there is no time to waste; at every second someone in the world is newly infected with TB bacilli (5). WHO has developed a Stop TB Strategy aimed at reducing the global burden of TB by 2015, by ensuring all TB patients access to high-quality diagnosis and patient-centered treatment. The strategy also supports the development of new and effective tools to prevent, detect and treat TB (5).
CONTRIBUTION OF BIOINFORMATICS TO THE RATIONAL DESIGN OF NEW DRUGS FOR TB
It is a fact that chemotherapy has lost potency against resistant strains of TB bacilli. Thus, TB treatment is entering a challenging era, where effective control requires the identification of new drugs and novel drug targets. Advances in this field have been recently made and some promising drug candidates like R207910 (79) and PA824 (80) are already in clinical trials (81). These drugs outperform the old ones because of their high activity against MDR strains and their potential to shorten the therapy.
In this field, bioinformatics may contribute to the rational design of new drugs and this will be later illustrated. Drug discovery in silico approaches requires structural 3D information either from the target or the ligand. These are classified in target-based virtual screening (TBVS) and ligand-based virtual screening based on the 3D primary information source being used (82). TBVS employs 3D structure of the protein target and ligand-based virtual screening uses known ligands 3D structures. Although, both methodologies are being used in TB drug design, only the results of applying TBVS are covered in this review. TBVS involves explicit molecular docking of each ligand into the binding site of the target, producing a predicted binding mode and a score for each database compound. Then, compounds are ranked according to their scores and only a small subset containing the top-ranking ones is selected for biological activity tests.
Many 3D structures of possible therapeutic targets for TB are available thanks to the efforts of the Tuberculosis Structural Genomics Consortium (TBSGC) (83), the Structural Proteomics in Europe (SpinE) consortium (84) and the XMTB Structural Proteomics Consortium (85). One goal of the TBSGC is to determine the structures of over 400 potential drug targets from the genome of M. tuberculosis and analyze their structures in the context of functional information. Selected protein targets belong to five different classes: extracellular proteins; iron-regulatory proteins; proteins targeted by known anti-TB drugs; proteins specific to mycobacteria and proteins with predicted novel folds. Some of these proteins were intentionally selected because of their participation in functional pathways related with pathogenesis, virulence and survival. Some others, especially those specific to mycobacteria whose functions are unknown, were involved in this study to gain further information on the above mentioned processes. About 229 different M. tuberculosis protein structures can be found in the Protein Data Bank (86), including 144 determined by TBSGC. These structural data have already been used in computerbased drug design. As result several lead compounds with the perspective to become anti-tuberculosis drugs have been identified, as shown below. They block proteins present in cellular pathways responsible for mycobacterial viability, pathogenicity and modulation of immune response, which constitutes successful targets for chemotherapy.
Computer-based inhibitor design for AccD5
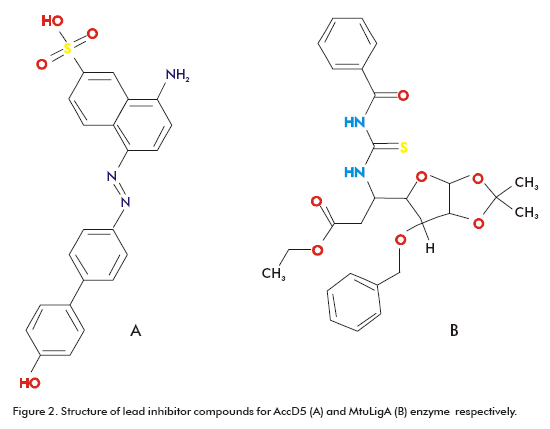
Some of these targets are proteins involved in cell envelope synthesis. Indeed, these are the targets for most of the antibiotics (e.g., isoniazid, ethionamide and ethambutol) used in chemotherapy. For example, one of the front-line antibiotics, isoniazid, targets the biosynthesis of mycolic acid, a major cell wall component that is unique to mycobacteria (87). This fatty acid is also important for antibiotic resistance, pathogen survival and virulence (88, 89). With the emergence of strains resistant to isoniazid, other targets in the biosynthesis pathways of cell wall lipids have been explored, such as AccD5. This enzyme has probed to be essential for methyl-branched long-chain acids biosynthesis (90). Bioinformatics has been applied in the search for drugs targeting AccD5. Specifically, the existing 3D structure of AccD5 (90) allowed the application of TBVS approach. As result, inhibitor compounds were identified from the National Cancer Institute (NCI) (91) diversity set and the University of California, Irvine, ChemDB database (92). The AccD5 inhibitor has a Ki of 13.1mM and binds AccD5 competitively with a 20-fold higher affinity than its substrates (Figure 2A) (90). The TBVS methodology was applied using the DOCK (93) and ICM-PRO (94) programs.
Computer-based inhibitor design for MtuLigA
Another target used in computer-based inhibitor design is a NAD-dependent DNA ligase, known as MtuLigA. This enzyme is essential in M. tuberculosis as showed by gene knockout experiments and other studies (95). M. tuberculosis codes for at least three different types of ATP-dependent ligases and a NAD-dependent ligase. The advantage of choosing LigA over ATP-dependent ligases is that the former exists only in bacteria and entomopox-viruses (96), in contrast to the second one, which is ubiquitously found (97). Srivastava et al. reported the crystal structure of the adenylation domain of MtuLigA bound to AMP (98). This crystal structure was used in TBVS using the programs AutoDock (99) and Gold (100). The 10% of the best scored ligands were selected for in vitro inhibitory activity evaluation assays. A novel class of specific inhibitors for MtuLigA was identified corresponding to glycosyl ureides, which were able to distinguish between NAD- and ATP- dependent ligases and inhibited MtuLigA in the micromolar range (Figure 2B) (98).
3D structural analysis of MbtLS complexes
TBVS involves several steps and it may become complex depending mostly on the selected docking program to be used. Sometimes, a simple but careful structural data analysis of protein 3D complexes may reveal new clues for drug design. Visualization and interpretation of the binding mode of inhibitors in their targets pocket in crystallographic complexes may provide a straight path to design novel inhibitors and to improve the potency of the already known ones. For example, the structural information obtained from the lumazine synthase (MbtLS) crystal structures with two inhibitor compounds 3-(1,3,7-trihydro-9-D-ribityl- 2,6,8-purinetrione-7-yl) propane 1-phosphate (TS-44) and 3-(1,3,7–trihydro-9-D-ribityl-2,6,8-purinetrion- 7-yl) butane 1-phosphate (TS-70) (101) demonstrated that compounds derived from purinetrione may serve as potential and specific inhibitors for MbtLS (Figure 3). MbtLS, like other enzymes involved in the biosynthesis of riboflavin, represent attractive targets for the development of drugs against bacterial pathogens, because the inhibition of these enzymes is not likely to interfere with enzymes of the mammalian metabolism. In particular, MbtLS catalyses the penultimate step of riboflavin biosynthesis pathway. Recently, new purinetrione compounds were identified based on the already known analogues mentioned before (101).
CONTRIBUTION OF BIOINFORMATICS TO THE UNDERSTANDING OF THE TB DISEASE
3D structural analysis of Rv1347c protein
Structural analysis has also been useful in addressing protein function linking with experimental studies, as is the case of the aminoglycoside N-acetyltransferase (Rv1347c) protein. Rv1347c was annotated as a possible aminoglycoside 6-N-acetyltransferase, which could not be demonstrated by biochemical assays (102). It was not until the resolution of its crystal structure and the corresponding structural analysis combined with functional data that its correct function was elucidated. In fact, Rv1347c belongs to the GCN5- related family of N-acyltransferases (GNAT), so it is not an aminoglycoside 6-N-acetyltransferase as it was previously suspected (103).
3D structural analysis of beta-lactamase
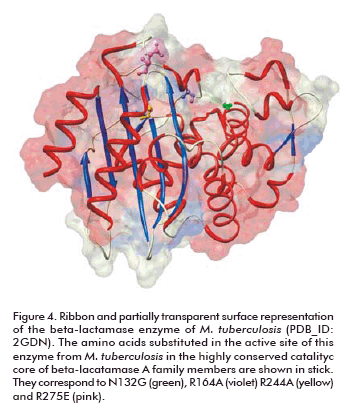
Beta-lactam antibiotics are extremely effective in disrupting the synthesis of the bacterial cell wall in both gram-positive and gram-negative bacteria, but not in M. tuberculosis. The enzyme responsible for beta-lactam resistance of TB is beta-lactamase. Thanks to the structural analysis of this enzyme, the features implicated in this resistance are now known at the atomic level. This helped in the understanding of the resistance mechanism and the design of drugs able to overcome resistance. The enzyme shares the common fold present in other class A beta-lactamases, but specific amino acids substitutions in the active site (N1 32G, R164A, R244A and R276E) account for its broad specificity, relatively low penicillinase activity and resistance to clavulanate (Figure 4) (104).
CONCLUSIONS
Undoubtedly, it is not simple to find a solution for TB and cholera but some improvements have been achieved and others are expected to come soon. In this sense, the exploitation of complete genome sequences and structural protein data through bioinformatic tools have allowed the rapid identification of new possible targets and drug candidates. In the case of TB, in silico approaches such as TBVS are being used in the design of novel drug candidates, some of them to overcome currently chemotherapy resistance. Although these drug candidates are still in a premature stage of development they may constitute the starting point to obtain better treatments for TB in a near future. In the case of cholera, other bioinformatics tools are being used such as those related to prediction of protein function, identification of hypothetical proteins and biochemical pathways analysis that might offer potential opportunities to improve the understanding about pathogenesis, virulence and resistance mechanism needed to discover new targets and efficient cholera vaccines.
REFERENCES
1. Satcher D. Emerging infections: getting ahead of the curve. Emerg Infect Dis 1995;1:1-6.
2. Global tuberculosis control: surveillance, planning, financing . World Health Organization Report 2006 (WHO/HTM/ TB/2006 362).
3. Morse SS. Factors in the emergence of infectious diseases. Emerg Infect Dis 1995;1:7-15.
4. Cholera annual report 2006. Weekly epidemiological record 2006; 2: 73-84.
5. Global tuberculosis control: surveillance, planning, financing . World Health Organization Report 2008 (WHO/HTM/ TB/2008 393).
6. Sack DA, Sack RB, Nair GB, Siddique AK. Cholera. Lancet 2004;363:223-33.
7. Barua D. History of cholera.. In: Cholera (Barua D and Greenough III WB, Eds) 1991, p. 1-35. Plenum, New York.
8. Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev 1998;62:1301-14.
9. Sarkar K. Role of oral rehydration therapy in controlling epidemic of cholera and watery diarrhoea. J Indian Med Assoc 2003;101:379-80,386.
10. Saha D, Karim MM, Khan WA, Ahmed S, Salam MA, Bennish ML. Single-dose azithromycin for the treatment of cholera in adults. N Engl J Med 2006;354:2452-62.
11. Das S, Saha R, Kaur IR. Trend of antibiotic resistance of Vibrio cholerae strains from East Delhi. Indian J Med Res 2008;127:478-82.
12. Roychowdhury A, Pan A, Dutta D, Mukhopadhyay AK, Ramamurthy T, Nandy RK, et al. Emergence of tetracyclineresistant Vibrio cholerae O1 serotype Inaba, in Kolkata, India. Jpn J Infect Dis 2008;61:128-9.
13. Griffith DC, Kelly-Hope LA, Miller MA. Review of reported cholera outbreaks worldwide, 1995-2005. Am J Trop Med Hyg 2006;75:973-7.
14. Levine MM, Black RE, Clements ML, Cisneros L, Nalin DR, Young CR. Duration of infection-derived immunity to cholera. J Infect Dis 1981;143:818-20.
15. Jertborn M, Svennerholm AM, Holmgren J. Evaluation of different immunization schedules for oral cholera B subunit-whole cell vaccine in Swedish volunteers. Vaccine 1993;11:1007-12.
16. Sanchez JL, Trofa AF, Taylor DN, Kuschner RA, DeFraites RF, Craig SC, et al. Safety and immunogenicity of the oral, whole cell/recombinant B subunit cholera vaccine in North American volunteers. J Infect Dis 1993;167:1446-9.
17. http://www.who.int/cholera/
18. Trach DD, Cam PD, Ke NT, Rao MR, Dinh D, Hang PV, et al. Investigations into the safety and immunogenicity of a killed oral cholera vaccine developed in Viet Nam. Bull World Health Organ 2002;80:2-8.
19. Trach DD, Clemens JD, Ke NT, Thuy HT, Son ND, Canh DG, et al. Field trial of a locally produced, killed, oral cholera vaccine in Vietnam. Lancet 1997;349:231-5.
20. Jertborn M, Svennerholm AM, Holmgren J. Intestinal and systemic immune responses in humans after oral immunization with a bivalent B subunit-O1/ O139 whole cell cholera vaccine. Vaccine 1996;14:1459-65.
21. Mahalanabis D, Lopez AL, Sur D, Deen J, Manna B, Kanungo S, et al. A randomized, placebo-controlled trial of the bivalent killed, whole-cell, oral cholera vaccine in adults and children in a cholera endemic area in Kolkata, India. PLoS ONE 2008;3:23-9.
22. Viret JF, Dietrich G and Favre D. Biosafety aspects of the recombinant live oral Vibrio cholerae vaccine strain CVD 103- HgR. Vaccine 2004;22:2457-69.
23. Richie EE, Punjabi NH, Sidharta YY, Peetosutan KK, Sukandar MM, Wasserman SS, et al. Efficacy trial of single-dose live oral cholera vaccine CVD 103-HgR in North Jakarta, Indonesia, a cholera endemic area. Vaccine 2000;18:2399-410.
24. Liang W, Wang S, Yu F, Zhang L, Qi G, Liu Y, et al. Construction and evaluation of a safe, live, oral Vibrio cholerae vaccine candidate, IEM108. Infect Immun 2003;71:5498-504.
25. Qadri F, Chowdhury MI, Faruque SM, Salam MA, Ahmed T, Begum YA, et al. Randomized, controlled study of the safety and immunogenicity of Peru-15, a live attenuated oral vaccine candidate for cholera, in adult volunteers in Bangladesh.J Infect Dis 2005;192:573-9.
26. García L, Jidy MD, Garcia H, Rodríguez BL, Fernandez R, Ano G, et al. The vaccine candidate Vibrio cholerae 638 is protective against cholera in healthy volunteers. Infect Immun 2005;73:3018-24.
27. Coster TS, Killeen KP, Waldor MK, Beattie DT, Spriggs DR, Kenner Jr, et al. Safety, immunogenicity, and efficacy of live attenuated Vibrio cholerae O139 vaccine prototype. Lancet 1995;345:949-52.
28. Larocque RC, Harris JB, Ryan ET, Qadri F, Calderwood SB. Postgenomic approaches to cholera vaccine development. Expert Rev Vaccines 2006;5:337-46.
29. Thiagarajah JR and Verkman AS. New drug targets for cholera therapy. Trends Pharmacol Sci 2005;26:172-5.
30. Reidl J, Klose KE. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol Rev 2002;26:125-39.
31. Fishmann PH. Mechanism of action of cholera toxin. In: ADP-ribosylating Toxins and G Proteins. (Moss J, Vaughan M, Eds).1990, 27-37. American Society for Microbiology, Washington DC.
32. Kaper JB, Fasano A, Trucksis M.. In: Vibrio cholerae and Cholera: Molecular to Global Perspectives (Wachsmuth KI, Blake PA and Olsvik O, Eds).1994, 45-76 American Society for Microbiology, Washington DC.
33. Islam MS, Miah MA, Hasan MK, Sack RB, Albert MJ. Detection of non-culturable Vibrio cholerae O1 associated with a cyanobacterium from an aquatic environmentin Bangladesh. Trans R Soc Trop Med Hyg 1994;88:298-9.
34. Yildiz FH and Schoolnik GK. Vibrio cholerae O1 El Tor: Identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA 1999;96:4028-33.
35. Watnick PI and Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol 1999;34:586-95.
36. Watnick PI, Lauriano CM, Klose KE, Croal L, Kolter R. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol Microbiol 2001;39:223-35.
37. Hammer BK and Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol 2003;50:101-4.
38. Lauriano CM, Ghosh C, Correa NE, Klose KE. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J Bacteriol 2004;186:4864-74.
39. Yildiz FH, Dolganov NA, Schoolnik GK. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J Bacteriol 2001;183:1716-26.
40. Casper-Lindley C and Yildiz FH. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J Bacteriol 2004;186:1574-8.
41. Waldor MK, Rubin EJ, Pearson GDN, Kimsey H, Mekalanos JJ. Regulation, replication, and integration functions of the Vibrio cholerae CTXphi are encoded by region RS2. Mol Microbiol 1997;24:917-26.
42. Kovach ME, Shaffer MD, Peterson KM. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology 1996;142:2165-74.
43. Waldor MK and Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 1996;272:1910-4.
44. Kirn TJ, Lafferty MJ, Sandoe CMP, Taylor RK. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol Microbiol 2000;35:896-910.
45. DiRita VJ, Parsot C, Jander G, Mekalanos JJ. Regulatory Cascade Controls Virulence in Vibrio cholerae. Proc Natl Acad Sci USA 1991;88:5403-7.
46. Higgins DE, DiRita VJ. Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol Microbiol 1994;14:17-29.
47. Hase CC and Mekalanos JJ. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA 1998;95:730-4.
48. Lee SH, Hava DL, Waldor MK, Camilli A. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 1999;99:625-34.
49. Lee SH, Butler SM, Camilli A. Selection for in vivo regulators of bacterial virulence. Proc Natl Acad Sci USA 2001;98:6889-94.
50. Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 2000;406:477-83.
51. Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V holerae. Lancet 2003;361:743-9.
52. Xu Q, Dziejman M, Mekalanos JJ. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc Natl Acad Sci USA 2003;100:1286-91.
53. Kim YR, Lee SE, Kim CM, Kim SY, Shin EK, Shin DH, et al. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect Immun 2003;71:5461-71.
54. Chen CY, Wu KM, Chang YC, Chang CH, Tsai HC, Liao TL, et al. Comparative Genome Analysis of Vibrio vulnificus, a Marine Pathogen. Genome Res 2003;13:2577-87.
55. Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci USA 2005;102:3004-9.
56. Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, et al. Host-induced epidemic spread of the cholera bacterium. Nature 2002;417:642-5.
57. Hommais F, Laurent-Winter C, Labas V, Krin E, Tendeng C, Soutourina O, et al. Effect of mild acid pH on the functioning of bacterial membranes in Vibrio cholerae. Proteomics 2002;2:571-9.
58. Kan B, Habibi H, Schmid M, Liang W, Wang R, Wang D, et al. Proteome comparison of Vibrio cholerae cultured in aerobic and anaerobic conditions. Proteomics 2004;4:3061-7.
59. Larocque RC, Krastins B, Harris JB, Lebrun LM, Parker KC, Chase M, et al. A Proteomic Analysis of Vibrio cholerae in Human Stool. Infect Immun 2008;76:4145-51.
60. Coelho A, de Oliveira SE, Faria ML, de Carvalho DP, Soares MR, von Kruger WM, et al. A proteome reference map for Vibrio cholerae El Tor. Proteomics 2004;4:1491- 504.
61. Kitano H. Systems biology: a brief overview. Science 2002;295:16624.
62. Onyebujoh P, Rook G. Tuberculosis. Nat Rev Microbiol 2004;2:930-2.
63. Zhang Y. Persistent and dormant tubercle bacilli and latent tuberculosis. Front Biosci 2004;9:1136-56.
64. OBrien RJ. Drug-resistant tuberculosis: etiology, management and prevention. Semin Respir Infect 1994;9:104-12.
65. Blumberg HM, Burman WJ, Chaisson RE, Daley CL, Etkind SC, Friedman LN, et al. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am J Respir Crit Care Med 2003;167:603-62.
66. Sierra GV. Is a new tuberculosis vaccine necessary and feasible? A Cuban opinion. Tuberculosis 2006;86:169-178.
67. Rodrigues LC, Diwan VK, Wheeler JG. Protective effect of BCG against tuberculous meningitis and miliary tuberculosis: a metaanalysis. Int J Epidemiol 1993;22:1154-8.
68. Brewer TF. Preventing tuberculosis with bacillus Calmette-Guerin vaccine:a metaanalysis of the literature. Clin Infect Dis 2000;31(Suppl 3):64-7.
69. Sterne JA, Rodrigues LC, Guedes IN. Does the efficacy of BCG decline with time since vaccination? Int J Tuberc Lung Dis 1998;2:200-7.
70. Sacksteder KA, Nacy CA. New tuberculosis vaccine development. Expert Opin Biol Ther 2002;2:741-9.
71. Brandt L, Feino CJ, Weinreich OA, Chilima B, Hirsch P, Appelberg R, et al. Failure of the Mycobacterium bovis BCG vaccine: some species of environmental mycobacteria block multiplication of BCG and induction of protective immunity to tuberculosis. Infect Immun 2002;70:672-8.
72. Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs-worldwide, 2000-2004. MMWR Morb Mortal Wkly Rep 2006;55:30-5.
73. Stewart GR, Robertson BD, Young DB. Tuberculosis: a problem with persistence. Nat Rev Microbiol 2003;1:97-105.
74. Horwitz MA, Lee BW, Dillon BJ, Harth G. Protective immunity against tuberculosis induced by vaccination with major extracellular proteins of Mycobacterium tuberculosis. Proc Natl Acad Sci USA 1995;92:1530-4.
75. Hess J, Miko D, Catic A, Lehmensiek V, Russell DG, Kaufmann SH. Mycobacterium bovis Bacille Calmette-Guerin strains secreting listeriolysin of Listeria monocytogenes. Proc Natl Acad Sci USA 1998;95:5299-304.
76. Horwitz MA. Recombinant BCG expressing Mycobacterium tuberculosis major extracellular proteins. Microbes Infect 2005;7:947-54.
77. Manabe YC, Scott CP, Bishai WR. Naturally attenuated, orally administered Mycobacterium microti as a tuberculosis vaccine is better than subcutaneous Mycobacterium bovis BCG. Infect Immun 2002;70:1566-70.
78. Mehta A, Tyagi RK, Goyal A, Khatri K, Gupta PNaVSP. Vaccination strategies for tuberculosis. Curr Sci 2007;93:1501-5.
79. Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 2005;307:223-7.
80. Stover CK, Warrener P, VanDevanter DR, Sherman DR, Arain TM, Langhorne MH, et al. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 2000;405:962-6.
81. Cole ST, Alzari PM. Towards new tuberculosis drugs. Biochem Soc Trans 2007;35:1321-4.
82. Lyne PD. Structure-based virtual screening: an overview. Drug Discov Today 2002;7:1047-55.
86. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res 2000;28:235-42.
87. Takayama K, Wang C, Besra GS. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin Microbiol Rev 2005;18:81-101.
88. Minnikin DE, Kremer L, Dover LG, Besra GS. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem Biol 2002;9:545-53.
89. Trivedi OA, Arora P, Vats A, Ansari MZ, Tickoo R, Sridharan V, et al. Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol Cell 2005;17:631-43.
90. Lin TW, Melgar MM, Kurth D, Swamidass SJ, Purdon J, Tseng T, et al. Structure-based inhibitor design of AccD5, an essential acylCoA carboxylase carboxyl-transferase domain of Mycobacterium tuberculosis. Proc Natl Acad Sci USA 2006;103:3072-7.
91. http://cactus.nci.nih.gov/ncidb2/
92. Chen J, Swamidass SJ, Dou Y, Bruand J, Baldi P. ChemDB: a public database of small molecules and related chemoinformatics resources. Bioinformatics 2005;21:4133-9.
93. Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des 2001;15:411-28.
94. Bursulaya BD, Totrov M, Abagyan R, Brooks CL, III. Comparative study of several algorithms for flexible ligand docking. J Comput Aided Mol Des 2003;17:755-63.
95. Gong C, Martins A, Bongiorno P, Glickman M, Shuman S. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J Biol Chem 2004;279:20594-606.
96. Sriskanda V, Moyer RW, Shuman S. NAD+ ependent DNA ligase encoded by a eukaryotic virus. J Biol Chem 2001;276:36100-9.
97. Wilkinson A, Day J, Bowater R. Bacterial DNA ligases. Mol Microbiol 2001;40:1241-8.
98. Srivastava SK, Tripathi RP, Ramachandran R. NAD+-dependent DNA Ligase (Rv3014c) from Mycobacterium tuberculosis. Crystal structure of the adenylation domain and identification of novel inhibitors. J Biol Chem 2005;280:30273-81.
99. Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated Docking Using a Lamarckian Genetic Algorithm and and Empirical Binding Free Energy Function. J Comput Chem 1998;19:1639-62.
100. Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved proteinligand docking using GOLD. Proteins 2003;52:609-23.
101. Morgunova E, Meining W, Illarionov B, Haase I, Jin G, Bacher A, et al. Crystal structure of lumazine synthase from Mycobacterium tuberculosis as a target for rational drug design: binding mode of a new class of purinetrione inhibitors. Biochemistry 2005;44:2746-58.
102. Draker KA, Boehr DD, Elowe NH, Noga TJ, Wright GD. Functional annotation of putative aminoglycoside antibiotic modifying proteins in Mycobacterium tuberculosis H37Rv. J Antibiot (Tokyo) 2003;56:135-42.
103. Card GL, Peterson NA, Smith CA, Rupp B, Schick BM, Baker EN. The crystal structure of Rv1347c, a putative antibiotic resistance protein from Mycobacterium tuberculosis, reveals a GCN5-related fold and suggests an alternative function in siderophore biosynthesis. J Biol Chem 2005;280:13978-86.
104. Wang F, Cassidy C, Sacchettini JC. Crystal structure and activity studies of the Mycobacterium tuberculosis beta-lactamase reveal its critical role in resistance to betalactam antibiotics. Antimicrob Agents Chemother 2006;50:2762-71.
Received in January, 2008.
Accepted for publication in March, 2009.
Arielis Rodríguez. Chemistry-Physics Division, Center for Genetic Engineering and Biotechnology, CIGB Ave 31 / 158 and 190, Cubanacan, Playa, PO Box 6162, Havana, Cuba. E-mail: arielis.rodriguez@cigb.edu.cu