Mi SciELO
Servicios Personalizados
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Biotecnología Aplicada
versión On-line ISSN 1027-2852
Biotecnol Apl vol.28 no.3 La Habana jul.-sep. 2011
RESEARCH
Low genetic variability in the fifth introduction of Litopenaeus vannamei in Cuba, as estimated with microsatellite markers
Limitada variabilidad genética de la quinta introducción en Cuba de Litopenaeus vannamei estimada con el uso de marcadores microsatélites
Adriana Artiles1, Ivón Rodríguez1, Anna Pérez2, Lourdes Pérez1, Georgina Espinosa2
1 Centro de Investigaciones Pesqueras. 5ta y 246, Barlovento, Santa Fé, Playa, La Habana, Cuba.
2 Facultad de Biología, Universidad de La Habana, UH. Calle 25 #455 e/ J y H, Vedado, Plaza, La Habana, Cuba.
ABSTRACT
The present work estimated the genetic variability and relatedness index of the fifth stock of Pacific white shrimp, Litopenaeus vannamei, imported into Cuba for farming purposes from the US Shrimp Improvement System (SIS). Genetic variability was estimated by genotyping 33 samples for four microsatellite loci: M1, Pvan 1815, Pvan 0040 and Pvan 1758. This stock had average expected and observed heterozygosities of 0.37 and 0.27 respectively; the lowest of all stocks previously introduced in Cuba. The above, together with the low amount of allelic variants detected for each microsatellite, was suggestive of low genetic variability. In addition, pairwise relatedness coefficients clustered around unity, indicating a high degree of consanguinity. Taken as a whole, the data suggests that this breeding stock should be crossed first with other individuals from a different source or with higher genetic variability.
Keywords: genetic variability, Litopenaeus vannamei, microsatellites, shrimp.
RESUMEN
Se estimó la variabilidad genética y el índice de parentesco entre lotes de camarón blanco del Pacífico, Litopenaeus vannamei, introducidos por quinta ocasión en Cuba, procedentes del Centro de mejora del camarón, de Estados Unidos (Shrimp Improving System: SIS), para su cultivo. La variabilidad genética se estimó mediante el genotipo de 33 muestras con cuatro loci microsatélites: M1, Pvan 1815, Pvan 0040 y Pvan 1758. El quinto lote de Litopenaeus vannamei tuvo los valores promedios de heterocigosidad esperada y observada, más bajos de todos los introducidos en Cuba: 0.37 y 0.27, respectivamente. Esto, unido a la poca cantidad de variantes alélicas para cada región microsatélite, indica una escasa variabilidad genética. Los valores del coeficiente de parentesco alrededor de la unidad expresan una alta consanguinidad. Todo ello sugiere el cruce de los individuos de esta introducción con otros de diferente origen o más variables genéticamente.
Palabras clave: variabilidad genética, Litopenaeus vannamei, microsatélites, camarón.
INTRODUCTION
Shrimp, as many other fishing resources, has received the impact of overfishing and environmental deterioration. Natural populations have shrunk, turning shrimp farming into a useful alternative for the obtention of this important protein source.
During the year 2003, two stocks of Pacific white shrimp (Litopenaeus vannamei) from the US Shrimp Improvement System (SIS), denominated here as scotcks 1 and 2, were introduced for the first time in Cuba [1], followed by the gradual implementation of techniques for the handling, health, nutrition and assessment of the genetic variability of this species in hatcheries and farms previously involved with Litopenaeus schmitti, an autochthonous shrimp. Two additional stocks were introduced during successive years (denominated here as stocks 3 and 4), all characterized using microsatellite markers [2, 3].
A fifth stock of this species, obtained from the same source, was introduced in October 2008. Its genetic composition and allele polymorphism are yet to be exa- mined, however. Although their consanguinity was low, the genetic variability of stocks 1 and 2 was already smaller than in natural populations [2]; stocks 3 and 4, likewise, exhibited a decreased genetic variability [3].
Using allozymes or microsatellites and a number of different shrimp species, it has been demonstrated that loss of genetic variability correlates with significant decreases in productivity (as for instance in Marsupenaeus japonicus [4], Litopenaeus stylirostris [5], P. monodon [6] and L. vannamei [7, 8]). The use of microsatellite markers, however, is better suited for the follow-up and assessment of genetic variability in farmed populations, due to its higher resolution and sensitivity [2, 9].
The objectives of the present work, therefore, are to determine genetic variability and relatedness indexes in a sample from the fifth stock of L. vannamei introduced in Cuba, employing four microsatellite loci and comparing them with all of the previously introduced stocks.
MATERIALS AND METHODS
Sampling
Pleopod samples were taken from the fourth pair, between the exopodito and the endopodito, of 40 randomly chosen individuals evenly split between males and females. The sampling procedure was performed on previously acclimatized shrimp from the hatcheries of the Shrimp Genetic Center in Mariel, Cuba.
Genotyping of microsatellite loci
DNA extraction procedures, microsatellite loci selected for analysis (M1, obtained by Wolfus et al. [8], Pvan 1758, Pvan 1815 and Pvan 0040, isolated by Cruz et al.[10]), amplification programs and electrophoresis conditions for genotyping runs have all been described in Borrell et al. [2].
DNA was extracted with 5% Chelex resin, and the amplified fragments were separated under a running voltage of 2000 mV in 6% bis-acrylamide/acrylamide gels which were later stained with 0.1% silver nitrate-0.05% formaldehyde, fixed with 10% acetic acid and developed with 3% sodium carbonate/0.05% formaldehyde and 20 µg/mL sodium thiosulfate. Samples previously genotyped by these authors [2] were used as controls for each microsatellite locus, with sizes ranging from 206 to 240 for M1, 140 to 146 for Pvan 0040, 110 to 136 for Pvan 1815 and 174 to 188 for Pvan 1758. PGEM® was also included as a conventional size marker.
Statistical processing
Determining the number of alleles per locus, the frequency of each allele and the values for observed (Ho) and expected (He, according to Nei [11]) heterozygosity for each locus, as well as whether the populations were in Hardy-Weinberg equilibrium, was performed with the GeneAlEx (version 6.1) software application [12]. Any locus with at least two alleles where the frequency of the most common allele did not exceed 95% was considered polymorphic [13].
Deviations from equilibrium were corroborated by calculating FIS [14], following the formula FIS = 1 - (Ho/He), with the FSTAT software application (version 2.9.3) [15]. Although it depends on population size, it is unaffected by the presence of multiple alleles per locus, the number of individuals per population or the number of populations.
The genetic relatedness coefficient (r) [16] for a single pair of individuals was also calculated with GeneAlEx ver. 6.1 [12]. This coefficient is calculated for codominant markers, using the following formula:
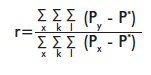
where: x stands for the individuals; k for the loci; l for allelic positions (two for diploids and one for haploids), Px is the frequency of individual “x” for locus k and allelic position l, Py is the frequency of allele “y” in the group or individual compared to x; and P* is the total frequency of the allele in the population. The genetic relatedness coefficient must be r ≤ 0 for non-related individuals; r = 0.25 for half brothers and r ≥ 0.5 for brothers [16].
RESULTS AND DISCUSSION
Genetic variability of the fifth stock of L. vannamei compared to previous stocks
The present work analyzed the genetic variability of the fifth stock of L. vannamei introduced in Cuba for shrimp farming, using four microsatellite loci: M1, isolated from L. vannamei [8], and Pvan 0040, Pvan 1758 and Pvan 1815, isolated from the same species [10].
The table contains the main calculated parameters: number of alleles (Na), observed and expected heterozygosity (Ho and He) and deviations from Hardy-Weinberg equilibrium (FIS), together with the values estimated during previous studies. According to the results of the analysis of linkage disequilibrium with FSTAT (version 2.9.3), these loci are not genetically linked in L. vannamei [2, 9, 17].
Expected and observed heterozygosity
Average observed heterozygosity for the fifth stock of L. vannamei introduced in Cuba yielded a value of 0.271; the lowest figure compared to previous introduced stocks (Figure 1). In addition, this value is below all previous intervals reported by other researchers employing microsatellites to study peneaid shrimps.
A now classic review [9] on the use of microsatellite loci for natural populations from four different species described observed heterozygosities that ranged from 0.425 to 0.964, yielding a mean of 0.666 which fell below the expected average (0.927). The same author observed heterozygosities ranging from 0.45 to 1.00 for three species of farmed peneaids, yielding a mean of 0.594, just below the mean for expected heterozygosity (0.674).
However, studies on P. stylirostris cultured for 22 and 24 generations [5] have produced much smaller values: Ho = 0.32 to 0.48; He = 0.46 to 0.61. Still, the values from the fifth stock introduced in Cuba (Ho = 0.271; He = 0.367) are below these figures (Figure 1), even though heterozygosity for all previous introductions of L. vannamei is within the above intervals. However, we agree that heterozygosity is an imperfect measure for variability, as it can yield high values with just two alleles, and, therefore, their quality also matters [17-19].
Allele frequencies and deviations from Hardy-Weinberg equilibrium
One single locus, Pvan 0040, is responsible to a large extent for the decrease in heterozygosity of the fifth stock, as it appears to be monomorphic in this case (as it did for the fourth Litopenaeus vannamei introduction). The other remaining three loci are still polymorphic, with five alleles each for M1 and Pvan 1758 and four alleles for Pvan 1815.
Allele frequencies for the loci of the fifth stock are shown in figure 2. Molecular weights for the observed alleles coincide within the intervals published by other authors, including those who first isolated them from a genomic library [10] and others who have later used them to characterize populations of this species [2, 3, 8, 17-19].
In the present work there were five M1 alleles ranging from 206 to 240 bp; the most frequent were those of 240 bp (0.61) and 210 bp (0.28). This microsatellite has previously been used in isolation to estimate genetic variability for different populations [8], obtaining allele numbers ranging from 4 to 23. This underscores the usefulness of this locus for population genetics studies in this species. M1 was also used for the genetic characterization of all previous introductions of L. vannamei into Cuba [2, 3].
The number of alleles per locus is also high for Pvan 1815 and Pvan 1758, both in natural populations and in farming environments subject to genetic improvement programs [10, 17-19]. Still, allelic variants are often lost in the latter, due to factors such as the consanguinity that typically appears in farmed populations and, probably, experimental artifacts such as the appearance of null alleles due to misinterpretation of polymerase slippages or the Wahlund effect, caused by the mixing of populations.
When these microsatellites were first obtained in 2002 [10] from a small sample, the observed numbers of alleles per locus were 12 and 14 for Pvan 1815 and Pvan 1758 respectively, suggesting they represented excellent markers. Later studies employing these microsatellites for studies of other L. vannamei populations have found similar allele numbers [17-19]. Their allele number in the present study, however, is lower (4 for Pvan 1815 and 5 for Pvan 1758) (Figure 2 and Table).
A retrospective look to previous stocks confirms that allele number has progressively decreased from earlier to later introductions for these and the other two loci. Finding out which specific alleles have been lost and determining the importance of these losses for future crossings or molecular marker-assisted selections would necessarily require, however, that all data be compared as a whole. As for locus Pvan 0040, which reappears here as a monomorphic allele and is one of the markers with the smallest number of allele variants. Although six alleles were found when it was first described [10], this study was only an initial report, based on a small sample. A later study from the same group where different populations were compared with the use of five microsatellite markers found 5 to 11 alleles for Pvan 0040 [17]. By the year 2009, however, this group had already stopped using this locus [19], employing Pvan 1758, Pvan 1815 and four new markers (Lvan 05 [20] and TUMXLv9.3, TUMXLv10.312 and TUMXLv8.256 [21]).
Using Pvan 0040 to characterize the two first stocks introduced in Cuba yielded 6 and 5 allele variants for stocks 1 and 2 [2], 5 alleles for the third, and, for the first time, only 1 allele for the fourth [3].
The results from the present work indicate that there is also a fixed Pvan 0040 allele for the fifth introduction of Litopenaeus vannamei in Cuba. Allele sizes in bp coincide between the fourth and the fifth introduction, despite the presence of disparities regarding sample sizes, methodologies for estimating genetic variability, reference molecular weight markers and positive controls. Future use of this marker for characterizing other stocks should, therefore, be reconsidered.
Although strict compliance with all conditions for reaching Hardy-Weinberg equilibrium is hard to come by in farmed populations, loci Pvan 1758 and Pvan 1815 do appear to be at equilibrium. M1 deviates from genetic equilibrium, however, mainly due to an excess of homozygotes as evidenced by the high value of FIS (Table). Previous studies [2, 3] of the introductions of L. vannamei in Cuba have evidenced significant deviations from genetic equilibrium, which in the presentwork take place only for M1. This phenomenon may be caused by a smaller sample size than that of other authors [2, 3], which would increase the probability of type I statistical errors; i.e. rejecting the null hypothesis when it is true.
Relatedness among the individuals of the fifth introduction of shrimps in Cuba
The analysis of the fifth introduction of L. vannamei shrimp in Cuba produced an anomalous curve with two maxima, corresponding to the region of highly related individuals (Figure 3). Note that the highest maximum corresponds to 1, that is, the highest possible degree of relatedness; the other maximum is at r = 0.15, close to that established by Queller and Goodnight [16] for half-brothers (r = 0.25). There is another peak to the left, although much smaller than the two others. Globally, average consanguinity yields a value of +0.037, implying that related individuals predominate in the sample.
The efficacy of the use of microsatellites and relatedness indexes to determine pedigrees was first shown while studying farmed turbot stocks with known consanguinities [22]. These authors obtained independent charts for related and non-related individuals, which implies that the frequency distribution of the relatedness indexes among individuals of the fifth introduction might actually correspond to the superimposition of these ideal curves. Most probably, the fifth introduced stock of L. vannamei actually constitutes a mixture.
Previous studies found genetic variability had decreased in stocks 3 and 4 of L. vannamei introduced in Cuba [3], but did not estimate genetic relatedness. However, since the first introductions, it became evident that the design of crossings using individuals from stock 2 (mean r = +0.1515) should not be undertaken without a methodology for following inter-individual genetic relatedness coefficients, which provide information on endogamy levels [2]. As mentioned above, average genetic relatedness in the present work is even higher, implying that these individuals should not be used as a breeding stock without an available technique for following up successive crossings.
Given the obstacles for the incorporation of individuals obtained from their natural environment into shrimp farming, the only available alternative for decreasing endogamy and raising genetic variability in L. vannamei stocks would be to measure these genetic parameters in the existing breeding stocks in Cuba, selecting the best according to all data.
CONCLUSIONS
Average expected and observed heterozygosity (0.37 and 0.27, respectively) of the fifth L. vannamei stock introduced in Cuba for shrimp farming were lower than those of previously imported stocks. This, together with the relatively small number of allele variants for each microsatellite region, indicates that its genetic variability is low. In addition, the fact that the genetic relatedness coefficient was close to unity indicates that its genetic consanguinity is high. According to the values obtained from the genetic parameters estimated in this work, it would therefore be necessary to cross this stock with others from the same source, in order to improve future production yields.
REFERENCES
1. Tizol R, Jaime B, Laria R, Pérez L, Machado R, Silveira R. Introducción en Cuba del camarón blanco del pacifico L. vannamei. Etapa I Cuarentena. Repositorio digital Ocean Docs; 2004 [cited 2010 Nov 16]. Available from: http://hdl.handle.net/1834/3588 .
2. Borrell YJ, Espinosa G, Vazquez E, Sánchez JA, Blanco G. Variabilidad genética de loci microsatélites en los primeros lotes de Litopenaeus vannamei introducidos en Cuba para la acuicultura. Rev Invest Mar. 2006;27(3):237-44.
3. Machado TR. Assessment of genetic variability in two lots of white shrimp, Litopenaeus vannamei (Boone, 1931) introduced to Cuba. International Fischeries Management [Master dissertation]. Department of Aquatic Biosciences. Norwegian College of Fishery Science: University of Tronso, Norway; 2006.
4. Sbordoni V, de Matthaeis M, Cobolli Sbordoni M, La Rosa G, Mattoccia M. Bottleneck effects and the depression of genetic variability in hatchery stocks of Litopenaeus japonicus (Crustacea, Decapoda). Aquaculture. 1986;57:239-51.
5. Bierne N, Beuzart I, Vonau V, Bonhomme F, Bedier E. Microsatellite-associated heterosis in hatchery-propagated stocks of the shrimp Peneaus stylirostris. Aquaculture. 2000;184:203-19.
6. Xu Z, Primavera JH, de la Pena LD, Pettit P, Belak J, Alcivar-Warren A. Genetic diversity of wild and cultured Black Tiger Shrimp (Penaeus monodon) in the Philippines using microsatellites. Aquaculture 2001;199:13-40.
7. Garcia DK, Faggart MA, Rhoades L, Alcivar-Warren AA, Wyban JA, Carr WH, et al. Genetic diversity of cultured Penaeus vannamei shrimp using three molecular genetic techniques. Mol Mar Biol Biotech. 1994;3:270-80.
8. Wolfus GM, García GK, Alcivar-Warren A. Application of the microsatellite technique for analyzing genetic diversity in shrimp breeding programs. Aquaculture. 1997;152:35-47.
9. Benzie JAH. Population genetics structure in peneaid prawns. Aquaculture Res. 2000;31:95-119.
10. Cruz P, Mejía-Ruiz CH, Pérez-Enriquez R, Ibarra AM. Isolation and characterization of microsatellites in Pacific white shrimp Peneaus (Litopenaeus vannamei). Mol Ecol Notes. 2002;2:239-41.
11. Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978;89:583-90.
12. Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288-95.
13. Graur D, W-H Li. Fundamentals of Molecular Evolution. 2nd ed. Sunderland (MA): Sinauer Asociates; 2000.
14. Wright S. The interpretation of population structure by F-statistics with special regards to systems of mating. Evolution. 1965;19:395-420.
15. Goudet J. FSTAT software (version 2.9.3.2). Lausanne: University of Lausanne; 2002 [updated 2005 Aug 23, cited 2010 Oct 14]. Available from: http://www.unil.ch/popgen/softwares/fstat.htm .
16. Queller DC, Goodnight KF. Estimating relatedness using genetic markers. Evolution. 1989;43(2):258-75.
17. Cruz P, Ibarra AM, Mejia-Ruiz H, Gaffney PM, Perez-Enríquez R. Genetic variability assessed by microsatellites in a breeding program of Pacific White Shrimp (Litopenaeus vannamei). Mar Biotechnol. 2004;6:157-64.
18. Valles-Jimenez R, Cruz P, Perez-Enriquez R. Population genetic structure of Pacific white shrimp (Litopenaeus vannamei) from Mexico to Panama: microsatellite DNA variation. Mar Biotechnol (NY). 2004;6(5):475-84.
19. Pérez-Enríquez R, Hernández-Martínez F, Cruz P. Genetic diversity status of White shrimp Penaeus (Litopenaeus vannamei) broodstock in Mexico. Aquaculture. 2009; 297:44-50.
20. Freitas PD, Jesús CM, Galleti PM Jr. Isolation and characterization of new microsatellite loci in the Pacific white shrimp Litopenaeus vannamei and cross-species amplification in other penaeid species. Mol Ecol Notes. 2007;7(2):324-6.
21. Meehan D, Xu Z, Zuniga G, Alcivar-Warren A. High frequency and large number of polymorphic microsatellites in cultured shrimp, Penaeus (Litopenaeus vannamei) [Crustacea:Decapoda]. Mar Biotechnol (NY). 2003;5(4):311-30.
22. Borrell YJ, Álvarez J, Vázquez E, Fernández Pato C, Martínez Tapia C, Sánchez JA, et al. Applying microsatellites to the management of farmed turbot stocks (Scophthalmus maximus L.) in hatcheries. Aquaculture. 2004;241:133-50.
Received in Decembre, 2010.
Accepted for publication in July, 2011.
Georgina Espinosa. Facultad de Biología, Universidad de La Habana, UH. Calle 25 #455 e/ J y H, Vedado, Plaza, La Habana, Cuba. E-mail: georgina@fbio.uh.cu .













