My SciELO
Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Biotecnología Aplicada
On-line version ISSN 1027-2852
Biotecnol Apl vol.28 no.4 La Habana Sept.-Dec. 2011
RESEARCH
Development, validation and application of a new ELISA for process control of the production of recombinant Hepatitis B surface antigen
Desarrollo, validación y aplicación de un nuevo ELISA para el control del proceso del antígeno de superficie el virus de la hepatitis B recombinante
Alberto Leyva, Julio C Sánchez, Lissette López, Milagros Font, Tatiana González, Bárbara Pérez, Neyda Hernández, Yamila Martínez, Annette Pereira, Ivonne Rodríguez
Centro de Ingeniería Genética y Biotecnología, CIGB. Ave. 31 entre 158 y 190, Cubanacán, Playa, CP 10 600, La Habana, Cuba.
ABSTRACT
The present work describes the development and validation of a sandwich-type enzyme-linked immunosorbent assay (ELISA) for quantifying the surface antigen of hepatitis B virus (HBsAg), obtained from a recombinant strain of the methylotrophic yeast Pichia pastoris. It is based on monoclonal antibody CB.Hep-1, normally employed as a ligand for the purification of HBsAg. Validation followed the guidelines of ICH Q2 and CECMED regulation No. 41 from 2007. Parameters such as linear working range, specificity, precision and accuracy were analyzed. The assay has a lower quantification limit of 11.9 ng/mL. Monoclonal antibody CB.Hep-1, used both for coating and as a conjugate during the ELISA, specifically bound recombinant HBsAg with excellent accuracy. An analysis of variance for the interference study yielded a probability, for each process control sample type/buffer combination, higher than 0.1, for a confidence level of 99%. Intra-assay variability ranged from 0.77 to 7.47%, and inter-assay variability ranged from 1.19 to 19.41%, always staying, for each sample, below 10 and 20% respectively. Recovery ranged from 98.18 to 100.31%, with a variation coefficient under 20%. The ELISA is specific for this monoclonal antibody within the range of studied concentrations, and has a linear response for antigen concentrations from 191.7 to 11.9 ng/mL. Given its precision, specificity and accuracy, this ELISA is a powerful tool for process control during the production of the recombinant vaccine against HBV.
Keywords: HBsAg, validation, Process control, ELISA.
RESUMEN
Se desarrolló y validó un ensayo inmunoenzimático en fase sólida (ELISA) de tipo sándwich para la cuantificación del antígeno de superficie del virus de la hepatitis B (VHB), obtenido por vía recombinante de la levadura metilotrófica de Pichia pastoris. La cuantificación se efectuó con el anticuerpo monoclonal (AcM) CB.Hep-1, utilizado como inmunoligando para su purificación. Los estudios de validación siguieron la guía ICH Q2 (R1) y la regulación No. 41, de 2007 del Cecmed. Se cuantificaron hasta 11.9 ng/mL, y se analizaron los parámetros de linealidad, especificidad, precisión y exactitud. El AcM CB.Hep-1, usado como recubrimiento y conjugado en el ELISA, fue específico en el reconocimiento del antígeno de superficie del VHB (AgsHB) recombinante, por lo que la cuantificación fue exacta. En el análisis de varianza para el ensayo de interferencia, el valor de probabilidad para cada tampón de las muestras del proceso de producción fue mayor que 0.1, para un nivel de confianza de 99%. La variabilidad intraensayo fue entre 0.77 y 7.47%, e interensayo fue entre 1.19 y 19.41%. Para cada muestra fue menor del 10 y el 20%, respectivamente. Se obtuvo un recobrado entre 98.18 y 100.31%, con un coeficiente de variación por debajo del 20%. El ELISA es específico para este anticuerpo monoclonal en el rango de concentraciones estudiadas. La curva de calibración es lineal entre 191.7 y 11.9 ng/mL. Por su precisión, especificidad y exactitud, este ELISA se convierte en una poderosa herramienta en el control del proceso de la vacuna recombinante contra el VHB.
Palabras clave: AgsHB, validación, control de procesos, ELISA.
INTRODUCTION
Hepatitis B still remains a worldwide health problem. According to current estimates, there are approximately 350 million carriers of the hepatitis B virus (HBV), which in addition causes, directly or indirectly, over half million deaths per year [1]. The use of recombinant DNA techniques has made possible the expression of the S gene for the surface antigen of the virus (HBsAg) in bacteria [2], yeast [3, 4], mammalian cells [5, 6], insect cells [7] and plant cells [8], allowing the large-scale obtention of this molecule for the production of an efficient vaccine against this pathogen.
HBsAg is a well characterized protein forming part of the HBV envelope. It self-assembles into very stable 24 nm subviral particles composed of one hundred or more monomers, held together by multiple intra- and inter-molecular disulfide bonds.
Since 1990, the Center for Genetic Engineering and Biotechnology (CIGB) at Havana has been producing HBsAg to be employed as the active pharmaceutical ingredient (API) of a vaccine against HBV, using an expression system based on the methylotrophic yeast Pichia pastoris. This vaccine is marketed un der the name Heberbiovac HB by the Cuban company Heberbiotec S. A. [9].
The process used to purify HBsAg at CIGB includes several steps, of which the most important is an immunoaffinity chromatography based on the interaction of HBsAg with the mouse monoclonal antibody (mAb) CB.Hep-1 [10-12]. This step removes most contaminating proteins with remarkable selectivity, yield and purity while also concentrating the sample significantly [13].
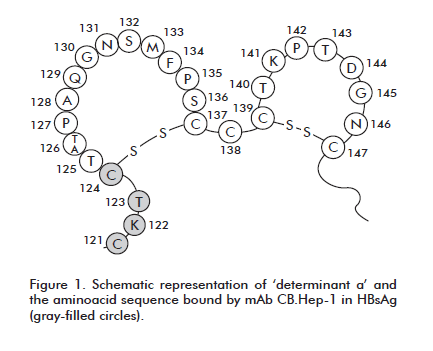
CB.Hep-1 binds an aminoacid sequence located at antigenic determinant ‘a’ of HBsAg [14]. This interaction, together with the multimeric nature of the viral particle, was exploited to develop and standardize a sandwich Enzyme-linked Immunosorbent Assay (ELISA) using solely this antibody both for capture and detection. The resulting assay is employed for quantifying HBsAg during its manufacturing process, from the stage of cell rupture to the obtention of the final purified API. However, this is an assay for content and potency [15, 16], which must therefore be validated according to requirements put in place in the biopharmaceutical industry to guarantee data reliability.
The present work describes the development, validation and application of a sandwich ELISA with mAb CB.Hep-1 for process control testing of the production of HBsAg. This assay allowed monitoring the yield of every purification stage based on the HBsAg concentration of the samples, with the accuracy and precision required by current international regulations.
MATERIALS AND METHODS
Enzymes and chemicals
All reagents and materials were purchased from Sigma (US), including the type VI-A horseradish peroxidase used for conjugation. They were employed following specifications from the manufacturers.
Biologicals
HBsAg (code: 11-03069), prepared by the Group of Stability and Reference Materials of the Quality Control Unit at CIGB, was employed as reference material. Monoclonal antibody CB.Hep-1 (code: 1AF0418), prepared and provided by the Monoclonal Antibodies Production Department at CIGB, was used as coating antibody. Peroxidase-linked CB.Hep-1 (code: Hep-1-4001) was obtained by the per-iodate method of Nakane and Kawaoi [17].
Samples and buffers from the production process
The samples used in the experiments were obtained from the production process for HBsAg. They were representative samples from each stage of the process: rupture supernatant (Rupt), supernatant from acid precipitation (SH), filtered Celite concentrate (CCT), non-bound fraction from the adjusted negative ion exchange (ION), immunoaffinity eluate (EIAF), eluate from 400 mM positive ion exchange (IIP) and active pharmaceutical ingredient (API). The buffers corresponding to each of these stages were: Rupt (20 mM Tris, 5 mM EDTA (w/v), 3 M KSCN, 0.3 M NaCl; pH 7), SH (20 mM Tris, 5 mM EDTA (w/v), 3 M KSCN, 0.3 M NaCl; pH 8), CCT (20 mM Tris, 3 mM EDTA, 250 mM NaCl), ION (20 mM Tris, 3 mM EDTA, 1 M NaCl), EIAF (20 mM Tris, 3 mM EDTA, 3 M KSCN, 1 M NaCl), IIP (20 mM Tris, 3 mM EDTA, 0.4 M NaCl; pH 7.2) and API (8 mM Na2HPO4 , 8 mM NaH2PO4 · H2O , 0.14 M NaCl; pH 6.7).
ELISA
One-hundred microliters of the CB.Hep-1 monoclonal antibody at 10 µg/mL in 0.05 M carbonate-bicarbonate coating buffer, pH 9.6 were used to coat each well of a MaxiSorp (Nunc) 96-well polystyrene plate for 20 min at 50 °C. A standard curve was prepared with serial dilutions of the reference material in the assay buffer (PBS 1X, 0.2%BSA, Tween 20 diluted ¼ 0.05%), and the control, together with the samples, were prepared at their working dilutions using the same buffer. They were added to the coated plate and incubated for 1 h at 37 °C. The plates were washed afterward, followed by the addition of 100 µL of the same antibody linked to horseradish peroxidase, diluted 1/20 000, leaving the mixture to react for 1 h a 37 °C on a water-saturated atmosphere. The reaction was developed using ortho-phenylenediamine (OPD) as substrate and 0.015% H2O2 in citrate buffer at pH 5.5. After 10 min color development was stopped by adding 50 µL of 2 M H2SO4, immediately reading the absorbance at 492 nm on a plate reader (Labsystem, Helsinki, Finland).
Determination of total protein concentration
The concentration of total proteins in process control samples from the production of HBsAg was determined by the Bradford method, using the modifications described by Zor and Selinger [18]. Samples, prepared at their established dilutions, were added into 96-well plates and immediately mixed with Coomassie reagent. After a 10 min incubation at room temperature, absorbance at 620/450 nm was determined on a plate reader (Labsystem, Helsinki, Finland). Total protein concentration was then determined by comparison against a standard curve of bovine serum albumin. The regression coefficient (r2) of the fitted standard curve had to be higher than 0.99 for the assay to be considered valid.
Experimental design
A response surface experimental design was used to optimize the concentration of mAb CB.Hep-1, time and incubation temperature for the coating step of the sandwich ELISA. The assayed values were transformed and codified as -1, 0 and 1 for statistical calculations. In this particular study the experimental design contains three treatments, including a central point and four replicates for each one. Data were fitted to a second order polynomial model, using the STATGRAPHICS centurion XV.1 (1994-2000) statistical software application. The assayed values were: 1, 5 and 10 µg/mL for CB.Hep-1 concentration, and 4 °C/14 hours, 37 °C/1 hour and 50 °C/20 minutes for incubation temperature/time.
Validation of the method
Linearity and working range
CB.Hep-1 concentrations ranging from 1.50 to 1500 ng/mL were used to evaluate assay performance with the reference material. The linear range of the curve was estimated using the least squares method, analyzing the regression coefficient (r2), the slope and the intercept of each curve.
Curve parameters (relationship between analyte concentration and the response, expressed as the absorbance at 492 nm) were determined with a standard curve prepared with the reference material at concentrations of 11.9, 23.9, 47.9, 95.8 and 191.7 ng/mL, with two replicates per point, and running a minimum of five independent assays. The working range was established as the widest range at which precision stayed beneath 20% and accuracy (recovery) stayed between 80-120%.
Specificity
Standard curves were prepared by dilution of the reference material either into assay buffer (PBS 1X, 0.2% BSA, 0.05% Tween 20) or into the different buffers forming part of process control samples from different purification stages, with the addition of the contaminants corresponding to each stage obtained from an HBsAg-negative control yeast strain. Care was taken to ensure that the concentration of buffers and contaminants corresponded to that produced by the usual working dilution of their corresponding sample. The obtained curves were compared using analysis of variance (ANOVA) with regard to their slope and intercept, also examining their regression coefficient (r2), the absorbance produced by blank samples and the concentration they yielded for the quantification of a positive control sample.
Accuracy
Accuracy was estimated by calculating recovery for each process control sample at three different concentrations of the reference material (95.8, 47.9 and 23.9 ng/mL), calculating Student’s t with a confidence of 95% for each sample. In addition, confidence intervals were estimated for the expected value.
Precision
The precision of the assay was estimated by comparing the results of independent determinations of the same samples from the production process. The coefficient of variation (CV) was calculated for three replicates of a single assay (repeatability), also examining the variability across three different days and three different analysts (intermediate precision). Reproducibility was studied by correlating an ELISA from the Quality Control (QC) unit with the validated assay from the Process Control (PC) laboratory, employing 15 determinations from different API batches for this purpose, and using, as acceptance criteria, a correlation coefficient (r) higher than 0.90.
RESULTS
Optimization of the assay
A sandwich-type ELISA was developed that is based on monoclonal antibody CB.Hep-1, used as an immunoligand in the purification of the hepatitis B surface antigen produced at the CIGB of Havana. This antibody binds the epitope known as ‘determinant a’, an immunodominant and protective region on the surface of the HBV virion (Figure 1).
The assay uses CB.Hep-1 for both capturing and detecting the antigen. A response surface-type experimental design was used for its optimization, in an effort to minimize the experimental work required for this purpose [19]. Tested variables included coating concentration and coating time and temperature, measuring their effect by following the final absorbance readings of the assay and evaluating their influence with a probability value. The ANOVA yielded a probability value higher than 0.05, indicating their effect was significant for a confidence level of 95%. According to the available data, the best results were obtained with a coating concentration of 10 µg/mL and an incubation of 20 minutes at 50°C. The equation for the model best fitting the data was:

Validation
Linearity and working range
The absorbances produced by a wide range of concentrations of the reference material (1.45 to 1550 ng/mL) were examined, using least squares fitting to examine their linearity (Figure 2A). Figure 2B shows the average of five independent standard curves, each prepared with the concentrations of the reference material spanning the linear range of the assay (191.7, 95.8, 43.5, 23.5 and 11.9 ng/mL). An ANOVA was used to analyze the results, which yielded an associated probability value lower than 0.01. This indicates a statistically significant relationship between concentration and absorbance for a 95% confidence level (Figure 2C). The regression coefficient (r2) indicates that a linear model, within this range, accounts for 98.39% of the variability of absorbance values. In all cases, blank samples yielded absorbances lower than 0.05, and the quantification limit was set at 11.9 ng/mL for a coefficient of variation of 8.6% (precision) and a recovery of 94% (accuracy).
Specificity
An ANOVA was used to compare the slopes and intercepts of standard curves of the reference material prepared either in assay buffer or in the buffer in which the different process control samples are received, with the addition of the yeast contaminants from the corres-ponding stage of the purification process, obtained from an HBsAg-negative recombinant yeast strain. Care was taken to ensure that the concentration of these buffers and contaminants matched that produced by the normal working dilution of the corresponding process control sample. The calculated probability according to the ANOVA was higher than 0.1 with a confidence of 99% (Table 1), and the parameters of each standard curve remained within the calculated limits, without statistically significant differences with the specifications established for this type of assay. The results demonstrate that the matrix in which the analyte is contained in process control samples does not interfere with its quantification by the CB.Hep-1-based ELISA.
Accuracy
Accuracy was estimated by calculating recovery at three different concentrations, according to CECMED regulation No. 41 from 2007 about the validation of analytical methods and regulation ICH Q2 (R1) from 2005 [15, 16]. In all cases recovery stayed within the established range of 80 to 120%, without statistically significant differences according to Student’s t-test. These results demonstrate that the assay is accurate, as the obtained values remained within the confidence interval calculated for each sample (Table 2).
Precision
Precision was determined with all process control samples employed in the validation of the assay. According to the repeatability study, the method has a variability lower than 10%. The results of the study of intermediate precision, per analyst and between analysts per sample type, are summarized in table 3. CV was lower than 20%.
The validation was completed with a joint reproducibility study involving the Quality Control unit of CIGB. It entailed the quantification of different API batches by the QC and PC laboratories, correlating the obtained results. Figure 3 plots the relationship between the QC and PC datasets corresponding to the CB.Hep-1 ELISA, and table 4 summarizes the results describing the relationship between both methods and setting the lineal model as control. The equation for the model was:
Given that the probability associated to F is lower than 0.01 for a 99% confidence, it is concluded that there is a statistically significant relationship between both methods.
The regression coefficient indicates that the chosen model accounts for the variability of the ELISA from the Process Control laboratory with respect to that of the Quality Control unit. The correlation coefficient was 0.9146, indicating a relatively strong relation between the examined variables. The standard error of the estimate exhibits a standard deviation for the residuals of 0.147; a value that can be used to build the limits for future observations. According to the results, there are no statistically significant differences pertaining the quantification of the API between both laboratories.
Application of the ELISA to the analysis of yields during the production process. Control chart
The performance and utility of the ELISA in real life conditions were evaluated by implementing it as part of the process control tests of the production of HBsAg.The analysis of yields of the process using this ELISA in combination with total protein determinations by the modified Bradford method [18] is shown in figure 4. This analysis was based on the data from five process batches, determining the coefficient of variation between yields of the same step for different batches; this CV was always lower than 25%, demonstrating the consistency of the process and the usefulness of the assay for adequately determining HBsAg concentrations in process control samples. In addition the assay was followed up uninterruptedly for a whole year, implementing a statistical control by means of a control chart used to evaluate the quantification of the positive control of the assay. A total of 200 determinations were performed; all of them fell within the established control limits, and the CV for all values was 8.84% (Figure 5). Only two values were separated from the central value by more than two standard deviations, representing 1% of the determinations. As a whole, the results demonstrate that both the control and the ELISA perform stably, and their quality is guaranteed on the long term.
DISCUSSION
The present work describes the development and validation of a new sandwich ELISA for the quantification of HBsAg in process control samples. Unlike similar reported assays [20], this one uses a single antibody. After optimization using a response surface methodology, an optimum was found at a coating concentration of 10 µg/mL and a coating time and temperature of 20 min at 50 ºC. The relatively high temperatures might favor the interaction of the antibody with the plastic surface, thus facilitating the capture of larger numbers of antigen molecules, as described by Leyva et al. [21].
Immunochemical techniques usually employ a standard curve to derive the value of unknown samples by interpolation [22]. In this study the standard curve was freshly prepared by serial dilutions of an HBsAg reference for each assay. The ELISA behaved linearly at antigen concentrations ranging from 191.7 to 11.9 ng/mL. This linear range is similar to that of similar ELISA, as that described by Karakuş et al. [23]. An important parameter to be examined during the validation of analytical methods, and especially so in immunoassays [22], is specificity, that is, their ability to unequivocally detect the analyte in the presence of the other components that will be present in the test mixture during real life use [15, 16]. In this case, the results of the interference tests demonstrated that the assay correctly measures the levels of HBsAg in the presence of all possible sample contaminants.
In order to determine whether the assay accurately quantifies the concentration of HBsAg in process control samples, samples of known concentration were assayed, then comparing the obtained and expected values. These experiments were done, for each sample type, at three different concentrations. In every case recovery varied from 98.18 to 100.31%, with an average of 100%. The results suggest, therefore, that the CB.Hep-1-based ELISA is appropriate for the quantification of HBsAg in biological samples.
Precision was evaluated by determining CV for two parameters: repeatability and intermediate precision, obtaining values lower than 10 and 20%, respectively. These values meet the acceptance criteria described in CECMED regulation No. 41 from 2007 and in IHC Q2 (R1) [15, 16], and do not reach the rejection threshold of 25% proposed by Findlay et al. for immunoenzyme assays [22], based on the fact that these techniques are less accurate than chromatographic methods and require larger number of experimental points to increase data reliability. The joint study between two laboratories demonstrated the reproducibility of the assay, with a correlation coefficient higher than 0.90 for the determinations performed in the submitted samples. This result coincides with those of Wapenaar et al. [24], who also obtained correlation coefficients with a value of 0.9 for a competition ELISA.
After validating an analytical method it is important to follow up and monitor its performance in actual op erating conditions. There are statistical techniques for the process control of recombinant proteins and the interpretation of results that constitute excellent tools for this purpose [25]. Control charts, in specific, represent the behavior of a specific parameter through time, making it easy to spot emerging trends and to assess the control, consistency and reliability of the underlying assay. The use of this technique with the present ELISA demonstrated the stability of the results, and its application to the analysis of process yields evidenced the consistency of the latter.
CONCLUSIONS
The validation of the ELISA for the quantification of HBsAg in process control samples indicates that the assay provides a linear response in the range of 191.7 to 11.9 μg/mL, with a regression coefficient higher than 0.98. The method was shown to be accurate, as there were no statistically significant differences between the known concentration of test samples and that determined with the assay; precision was acceptable, with a variability lower than 10% for the repeatability study, and lower than 20% for the study of intermediate precision. There is a strong correlation between the concentrations determined at the Quality Control and Process Control laboratories, performed during the reproducibility study. The method is adequate and provides reliable data on the behavior of the purification process for HBsAg. After comparing the data with established acceptance criteria, it is concluded that the validation is satisfactory and, therefore, the assay can be used for process control of the production of HBsAg.
REFERENCES
1. Centers for Disease Control and Prevention. Achievements in public health: hepatitis B vaccination. United States, 1982-2002. MMWR Morb Mortal Wkly Rep. 2002;51(2):529-52.
2. Burrell CJ, Mackay P, Greenaway PJ, Hofschneider PH, Murray K. Expression in Escherichia coli of hepatitis B virus DNA sequences cloned in plasmid pBR322. Nature. 1979;279(5708):43-7.
3. Miyanohara A, Toh-e A, Nozaki C, Hamada F, Ohtomo N, Matsubara K. Expression of hepatitis B surface antigen gene in yeast. Proc Natl Acad Sci USA. 1983;80(1):1-5.
4. Gellissen G, Janowicz ZA, Weydemann U, Melber K, Strasser AW, Hollenberg CP. High-level expression of foreign genes in Hansenula polymorpha. Biotechnol Adv. 1992;10(2):179-89.
5. Hsiung N, Fitts R, Wilson S, Milne A, Hamer D. Efficient production of hepatitis B surface antigen using a bovine papilloma virus-metallothionein vector. J Mol Appl Genet. 1984;2(5):497-506.
6. Holzer GW, Mayrhofer J, Leitner J, Blum M, Webersinke G, Heuritsch S, et al. Overexpression of hepatitis B virus surface antigens including the preS1 region in a serum-free Chinese hamster ovary cell line. Protein Expr Purif. 2003;29(1):58-69.
7. Kang CY, Bishop DH, Seo JS, Matsuura Y, Choe M. Secretion of particles of hepatitis B surface antigen from insect cells using a baculovirus vector. J Gen Virol. 1987;68(Pt 10):2607-13.
8. Mason HS, Ball JM, Shi JJ, Jiang X, Estes MK, Arntzen CJ. Expression of Norwalk virus capsid protein in transgenic tobacco and potato and its oral immunogenicity in mice. Proc Natl Acad Sci USA. 1996;93(11):5335-40.
9. Muzio VL, Pentón E, Palou M, Fontirrochi G, Nazábal M, González MJ, et al, inventors; Centro de Ingeniería Genética y Biotecnología, assignee. Method for obtaining recombinant surface antigen of hepatitis B virus (HEP B) of higher immunogenic capacity and use thereof in a vaccine preparation. European Patent EP480525. 1992 Apr 15.
10. Agraz A, Duarte CA, Costa L, Pérez L, Páez R, Pujol V, et al. Immunoaffinity purification of recombinant hepatitis B surface antigen from yeast using a monoclonal antibody. J Chromatogr A. 1994;672(1-2):25-33.
11. Páez R, Agraz A, Herrera L. The recovery of the hepatitis B virus surface antigen (AgsHB) from a recombinant P. pastoris strain, disruption and precipitation studies. Acta Biotechnol. 1993;13:117-22.
12. Pérez L, López S, Beldarraín A, Arenal D, Pentón E. Purification of recombinant hepatitis B surface antigen (rec-HBsAg) from P. pastoris: a process development study. In: Galindo E, Ramírez OT, editors. Advances in Bioprocess Engineering. Dordrecht: Kluwer Academic Publishers; 1994. p. 527-34.
13. Hardy E, Martínez E, Diago D, Díaz R, González D, Herrera L. Large-scale production of recombinant hepatitis B surface antigen from Pichia pastoris. Biotechnol. 2000;77(2-3):157-67.
14. Fernández de Cossío ME, Díaz T, Galván A, Valdéz R, González E, Ayala M, et al. Antigen recognition characteristics and comparative performance in immunoaffinity purification of two monoclonal antibodies specific for the hepatitis B virus surface antigen. J Biotechnol. 1997;56(2):69-80.
15. Centro para el Control Estatal de la Calidad de los Medicamentos (CECMED). Regulación No. 41-2007: Validación de Métodos Analíticos. Ámbito Regulador. 2007 Mar 3;(53):1-14.
16. International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use. Validation of Analytical Procedures: Text and Methodology Q2(R1). ICH; 2005 Nov.
17. Nakane PK, Kawaoi A. Peroxidase-labeled antibody. A new method of conjugation. J Histochem Cytochem. 1974;22(12):1084-91.
18. Zor T, Selinger Z. Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies. Anal Biochem. 1996;236(2):302-8.
19. Myers RH, Montomery DC. Response surface methodology: process and process optimization using designed experiments. 2nd ed. NewYork: John Wiley & Sons; 2002.
20. van Roosmalen MH, de Jong JJ, Haenen W, Jacobs T, Couwenberg F, Ahlers-de Boer GJCM, et al. A new HBsAg screening assay designed for sensitive detection of HBsAg subtypes and variants. Intervirology. 2006;49:127-32.
21. Leyva A, Franco A, González T, Sánchez JC, López I, Geada D, et al. A rapid and sensitive ELISA to quantify an HBsAg specific monoclonal antibody and a plant-derived antibody during their downstream purification process. Biologicals. 2007;35(1):19-25. 22. Findlay JW, Smith WC, Lee JW, Nordblom GD, Das I, DeSilva BS, et al. Validation of immunoassays for bioanalysis: a pharmaceutical industry perspective. J Pharm Biomed Anal. 2000;21(6):1249-73.
23. Karakuş R, Aral LA, Baştürk B, Aybay C. Development of a highly sensitive ELISA for quantification of hepatitis B virus (HBV) surface antigen (HBsAg). Turk J Med Sci. 2007;37(2):87-92.
24. Wapenaar W, Barkema HW, Vanleeuwen JA, McClure JT, O’Handley RM, Kwok OC, et al. Comparison of serological methods for the diagnosis of Neospora caninum infection in cattle. Vet Parasitol. 2007;143(2):166-73.
25. Fuge J. FDA CONFERENCE Report: Validation Considerations When Preparing For FDA Inspections. J Validat Technol. 2005;11(3):253-6.
Received in December, 2010.
Accepted for publication in September 2011.
Alberto Leyva. Centro de Ingeniería Genética y Biotecnología, CIGB. Ave. 31 entre 158 y 190, Cubanacán, Playa, CP 10 600, La Habana, Cuba. E-mail: alberto.leyva@cigb.edu.cu.