Mi SciELO
Servicios Personalizados
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Biotecnología Aplicada
versión On-line ISSN 1027-2852
Biotecnol Apl vol.30 no.1 La Habana ene.-mar. 2013
RESEARCH
Molecular identification of thirteen isolates of Trichoderma spp. and evaluation of their pathogenicity towards Rhizoctonia solani Kühn
Identificación molecular y evaluación patogénica de trece aislamientos de Trichoderma spp. frente a Rhizoctonia solani Kühn
Danay Infante1, Benedicto Martínez1, Belkis Peteira1, Yusimy Reyes2, Alfredo Herrera3
1 Grupo de Fitopatología, Dirección de Protección de Plantas, Centro Nacional de Sanidad Agropecuaria, Censa. Autopista Nacional km 23½, AP 10, San José de las Lajas, Mayabeque, Cuba.
2 Facultad de Agronomía, Universidad Agraria de La Habana Fructuoso Rodríguez Pérez, UNAH Autopista Nacional km 23½, AP 10, San José de las Lajas, Mayabeque, Cuba.
3 Laboratorio Nacional de Genómica para la Biodiversidad, Langebio, Centro de Investigación y de Estudios Avanzados, Cinvestav, Instituto Politécnico Nacional, IPN. CP 36821, Irapuato, Guanajuato, México.
ABSTRACT
Disease caused by Rhizoctonia solani Kühn infestations has become a growing problem for commercially important crops. Although this fungus is usually controlled through the application of chemicals, the heavy ecological and financial toll of the latter has prompted for research on biopesticides as a viable alternative. Trichoderma spp. is a well-known fungus often used for the biological control of crop pests, whose anti-fungal mechanisms include competition for the substrate, antibiosis and/or mycoparasitism. In the present work, we have used molecular techniques (sequencing of amplicons from the internal transcribed spacer of ribosomal DNA and the EF1A translation elongation factor) for the taxonomic identification of 13 Trichoderma spp. isolates in our collection, also evaluating their antibiotic effect on strains from three anastomosis groups of R. solani (AG-2.1, AG-5, AG-8) by the cellophane method. The sequences obtained from all isolates exhibited 100 % identity with deposited T. asperellum Samuels sequences in TrichoBLAST/GenBank, enabling their taxonomic assignment to this species. When analyzed by in vitro tests, over 70 % of the isolates exhibited a fungistatic effect towards R. solani, with the remaining strains exhibiting fungicidal activity; these results were later corroborated by technical efficacy tests under field conditions. Isolates 3, 13, 17, 75, 78, 85 and 90 were selected as potential biocontrol agents due to their high antibiotic activity and technical efficacy under field conditions.
Keywords: Trichoderma asperellum, Rhizoctonia solani, internal transcribed spacer, transcriptional elongation factors, fungicide effect, fungistatic effect.
RESUMEN
La incidencia de las enfermedades causadas por Rhizoctonia solani Kühn en las plantas aumenta cada año. Su control es fundamentalmente con productos químicos, en su mayoría muy tóxicos. Por tal razón, se investigan alternativas bioplaguicidas. Trichoderma spp. es uno de los hongos más utilizados para el control biológico de plagas, por sus propiedades y mecanismos de acción: competencia por el sustrato, antibiosis y micoparasitismo. Este trabajo tuvo como objetivos la identifi cación molecular y evaluación patogénica de 13 aislamientos del género Trichoderma frente a aislamientos de R. solani. Se evaluó su efecto antibiótico sobre aislamientos de 3 grupos de anastomosis de R. solani (AG-2.1, AG-5, AG-8) por el método de celofán. La identificación molecular partió de la secuenciación del espaciador interno del transcrito de la región del ADN ribosomal y del factor de elongación de la traducción EF1A. Más del 70 % de los aislamientos evaluados ejerció un efecto fungistático; mientras que el resto actuó como fungicida. La evaluación de la eficacia técnica en condiciones de campo corroboró estos resultados. A partir de la secuenciación y comparación de los datos de las secuencias depositadas en TrichoBLAST/GenBank, se comprobó que todos los aislamientos tenían el 100 % de identidad con la especie Trichoderma asperellum Samuels. Se seleccionaron los aislamientos 3, 13, 17, 75, 78, 85 y 90 como promisorios agentes de control biológico, por su alto potencial antibiótico y eficacia técnica en condiciones de campo.
Palabras clave: Trichoderma asperellum, Rhizoctonia solani, espaciador interno del transcrito, factores de elongación transcripcional, efecto fungicida, efecto fungistático.
INTRODUCTION
Plant pathogens and their antagonist species exist in the context of a complex web of ecological interactions. Fungi such as Trichoderma harzianum Rifai, T. auroviride Rifai and T. asperellum Samuels, for instance, have been shown to play an important regulatory role [1], counteracting a number of fungal plant pathogens through antibiosis, mycoparasitism or simply competing for their substrate. Not surprisingly, Trichoderma has long been a subject of study for its potential application in the field of biological control of phytopathogens [2-4]. Trichoderma, a member of the Hypocrea family, is responsible for the formation of a considerable part of soil biomass [5], participates in mutualistic associations with plant roots which increase considerably their nutrient uptake capabilities [6], and induces systemic resistance in their hosts to a number of phytopathogens [7-10].
According to Samuels [11] and the Crop Protection Compendium [12], the teleomorphs of some Trichoderma species are typical of the Hypocrea genus (such as Ascomycetes and Hypocreales). Rifai [13] used morphological traits (conidiophore branching patterns and conidial shape) to provide a first approximation to the diversity and structure of Trichoderma spp., describing nine species (T. hamatum (Bon.) Bain, T. viride Pers. ex S. F. Gray, T. aureoviride, T. harzianum, T. koningii Rifai, T. pseudokoningii Rifai, T. longibrachiatum Rifai, T. polysporum (Link ex Pers) Rifai and T. piluliferum Webster & Rifai) as an aggregate species and pointing out that T. hamatum might be formed by two or more morphological distinct species. To compound matters, these species are in constant variation, and it has therefore been impossible to provide an accurate description of their characteristics, even with molecular methods [14]. Bissett et al. [15-17] later described the Longibrachiatum genus section, grouping T. viride, T. koningii, T. pseudokoningii, T. longibrachiatum, T. citrinoviride Bissett and T. atroviride Bissett together.
The research of Samuels et al. [18, 19] on the Hypocrea schweinitzii Samuels and Trichoderma sect. Longibrachiatum complex provided new data regarding the taxonomy of hitherto unknown species of the genus. Samuels et al. [19] and Lieckfeldt et al. [20] examined T. viride, a species initially defined solely by the presence of conidial warts, concluding that there were two morphologically distinct types (I and II) with differences in mitochondrial DNA. Later morphological, physiological and molecular studies established that morphological type I corresponded to the actual T. viride species (anamorph of Hypocrea), whereas type II actually corresponded to a new species of T. asperellum, characterized molecularly and close to the T. hamatum neotype.
Several T. asperellum isolates are currently used as biopesticides for the control of pathogenic fungi in economically important crops, both under greenhouse and open field conditions. Their mycoparasitic effect is highly specific for Rhizoctonia solani and Sclerotium spp., and is based on the production of high levels of chitinolytic enzymes [1, 9, 18, 21]. The Plant Mycology Laboratory of the National Center for Agricultural Health (Censa, Cuba) has put together a strain collection conserving over 80 Trichoderma spp. isolates, among which there is a promising group of indigenous isolates with morphocultural and pathogenic traits distinct from those of T. harzianum strains A 34, A 53 and Ts-3, currently employed for the production of biopesticides. The present work, therefore, centers on the molecular characterization of 13 of these isolates of the Trichoderma genus and the evaluation of their pathogenicity towards isolates of R. solani.
MATERIALS AND METHODS
Trichoderma spp. isolates
This study used 14 samples of T. atroviride (IMI206040) as reference strain for molecular tests, and isolates T.1, T.3, T.12, T.13, T.17, T.25, T.28, T.56, T.75, T.78, T.79, T.85 and T.90 from the Plant Mycology Laboratory of Censa, Cuba.
Rhizoctonia isolates
This study used isolates from three anastomosis groups of R. solani (AG-2.1, AG-5, AG-8), provided by the National Genomics Laboratory for Biodiversity (Langebio) of the Center for Advanced Research and Studies (Cinvestav) of the National Polytechnic Institute (IPN), México.
Culture conditions
The Trichoderma spp. and R. solani isolates were seeded on potato-dextrose agar medium (APD, Difco) in 90 mm Petri dishes and incubated on total darkness at 28 ± 2 °C and 25 ± 2 °C, respectively. When fungal growth covered almost the entire surface of the plate, 6 mm disc samples were taken from the periphery of the colonies for further experimentation.
In vitro antibiotic effect of Trichoderma spp. isolates towards isolates from three anastomosis groups of R. solani
Samples of Trichoderma spp. (excepting T. atroviride, IMI206040) were seeded on 90 mm Petri dishes with PDA medium (Difco) covered with a cellophane membrane [22] by placing the individual mycelial discs on the center of the plate (non-inoculated controls were also set up), incubating the plates for 36 h at 28 ± 2 °C in absolute darkness until fungal growth reached ¾ of plate diameter. At this point the cellophane membrane was retired, and the plates were then inoculated separately with isolates from the different R. solani anastomosis groups and incubated under the conditions described above for this phytopathogen. Three replicates were used per isolate, applying a random design. Radial fungal growth was measured at 24, 48 and 72 h post-inoculation, using the inhibition of radial growth (ICR) as response variable:

where:
R1: radial growth of the R. solani sample in the control plate without Trichoderma sp. (mm);
R2: radial growth of the R. solani test sample (mm).
Extraction and purification of genomic DNA
A cellophane-covered PDA (Difco) plate was inoculated by placing one disc of the relevant Trichoderma spp. isolate in the center of the dish, incubating the culture for 3 days at 28 ± 2 °C. The mycelium was then collected into aluminum foil with the help of a spatula and stored at -20 °C. Genomic DNA was extracted by the urea-phenol-chloroform method standardized at Langebio, Mexico. Namely, 600 µL of extraction buffer (42.04 % urea, 0.3 M NaCl, 0.02 M EDTA, 0.05 M Tris-HCl, pH 8) were added to 100 ng of mycelium, homogenizing the resulting mixture and incubating it for 30 min. at room temperature. Then, 300 µL of phenol-chloroform (25:25 v/v) were added and the mixture was homogenized for 10 min by vortexing, and centrifuged afterwards for 15 min at 10 000 rpm in a microcentrifuge. The supernatant was collected and the extraction with phenol-chloroform was repeated as described above. Two-hundred microliters of isopropanol were added to the resulting supernatant, mixed and incubated overnight at -20 °C, collecting the precipitated DNA by centrifugation for 5 min at 10 000 rpm. The pellet was rinsed with 200 µL of 70 % EtOH and resuspended into 40-60 µL of sterile distilled water. The quality of the purified DNA samples was checked by electrophoresis in 1.5 % agarose gels using TAE 1× as running buffer, for 45 min at 100 V.
Amplification by Polymerase Chain Reaction of the internal spacer of rDNA and elongation factor
In order to amplify fragments of the internal transcribed spacer of 5.8 S rDNA (ITS1 and ITS2), DNA samples were subjected to amplification by PCR in a total volume of 50 µL containing 1× magnesium-free PCR buffer, MgCl2 1.5 mM; dNTP 0.4 mM; 2.5 U of recombinant Taq DNA polymerase; 100 ng of total genomic DNA and 0.2 μM of each primer. Two primer pairs were used to amplify the target: either ITS1 (5’ TCC GTA GGT GAA CCT GCG G 3’) and ITS2 (5’ GCT GCG TTC TTC ATC GAT GC 3’) or ITS3 (5’ GCA TCG ATG AAG AAC GCA GC 3’) and ITS4 (5’ TCC TCC GCT TAT TGA TAT GC 3’) [23].
A 0.4 kb fragment from the gene coding for translational elongation factor (Tef) eEFa1, containing three introns, was also amplified as described above but in a volume of 25 µL and using primers tef 1a fw (5´ CTA CGA GAA GTT CGA GAA GG 3´) and tef 1a rev (5´ TAC TTG AAG GAA CCC TTA CC 3´) [23], each at 0.2 μM.
The amplification reactions were performed in an API thermal cycler (Applied Biosystems, USA), using an initial denaturation step of 94 °C for 3 min, followed by 30 cycles of a 40 s denaturation step at 94 °C, a 40 s annealing step at 50 °C and a 90 s extension step at 72 °C, followed by a single final extension step of 7 min at 72 °C (Tef reactions used instead an annealing temperature of 56 °C). Five-microliter samples of each reaction were analyzed by electrophoresis in 1 % agarose gels using TAE 1× (Tris base 4.84 g, acetic acid 1.142 g, Na2 EDTA 2 H2O 0.74 g) as running buffer at 100 V for 15-20 min followed by visualization in a gel documentation workstation. The size of the resulting amplicons was estimated by comparison to the relative migration of a 100 bp DNA sizing ladder (Invitrogen, USA).
DNA sequencing
A spin column was used to purify 25 µL of each PCR amplification reaction, which were then sent to the core sequencing facilities of Langebio (Cinvestav, México). The isolates were identified by comparing the resulting sequences against those of TrichoBLAST/GenBank [24].
Efficacy of the action of T. asperellum isolates on R. solani under field conditions
Fungal inocula were obtained by seeding pure isolates of T. asperellum (excepting isolate T.56) and R. solani into malt-agar and PDA plates, respectively (National Center for Biopreparations, Biocen, Cuba), which were then incubated at 28 ± 2 °C and 25 ± 2 °C respectively under total darkness until fungal growth covered the entire surface of the plate. The experiment was performed on parcels of pH 5.4 red ferrallitic soil, set aside for over 2 years, without weeds, kept moist throughout the experiment. Parcels with an area of 1 m2 were demarcated, spaced 50 cm apart, and four spots were marked on each one. Then, 50 mL of a suspension containing 104 sclerotia/mL of the pathogenic microorganism to be tested were poured on each of the four marked spots of each parcel. After 21 days, each of these spots was then inoculated with 50 mL of a suspension of the corresponding Trichoderma spp. isolate at 107 conidia/mL. One-kilogram soil samples from each spot were then taken after 15 days and processed independently. After homogenizing and sieving the samples, 1 g portions were subjected to serial 10-3 dilutions, inoculating 10 µL of each dilution uniformly into PDA plates (Biocen, Cuba) that were then incubated at 28 ± 2 °C under total darkness. Five replicates were set up per treatment, including a control inoculated solely with the pathogen. After 72 h, the number of R. solani colonies per plate and per gram of soil was estimated, calculating treatment efficacy for each Trichoderma spp. isolate with Abbott’s formula [25].
RESULTS AND DISCUSSION
In vitro antibiotic effect of thirteen Trichoderma spp. isolates against isolates from three anastomosis groups of R. solani
The metabolites excreted by 11 of the studied Trichoderma spp. isolates inhibited growth of the Rhizoctonia isolate from anastomosis group AG-2.1 by 60 to 80 %, in contrast with the remaining two isolates (T.75 and T.90), which were fungicidal (Figure 1). All Trichoderma spp. isolates inhibited growth of the R. solani isolate from anastomosis group AG-5 (one of the most aggressive rice phytopathogens) by 50 to 90 %, but none were fungicidal. In the case of the Rhizoctonia isolate from anastomosis group AG-8, the metabolites from Trichoderma isolates T.3, T.12, T.17, T.28, T.56, T.75 T.78 and T.79 were fungicidal, and the remaining isolates inhibited growth of the pathogen by 70 to 80 %. These activities are mediated by the secretion of volatile and non-volatile secondary metabolites by many Trichoderma spp. strains, which diffuse out and inhibit the growth of other microorganisms, exhibiting therefore a fungistatic effect (these compounds are commonly considered as ‘antibiotics’) that does not require direct contact [1].
Küçük and Kivanç [26] previously obtained similar results with the cellophane method. In their case, the growth inhibition of Gibberella zeae (Schwein) Petch was larger than 60 %, and the metabolites from isolates Tm4 and Tm9 inhibited the growth of Aspergillus ustus (Bain) Thom & Church by 50 %. Similar results were reported for isolates Tm7 and Tm10 against T. harzianum.
On the other hand, Infante [27] as well as Martínez et al. [28] produced growth inhibitions larger than 50 % for R. solani when using the dual culture method with two isolates of Trichoderma spp. It should be noticed, however, that Trichoderma spp. can excrete in both cases (dual culture and cellophane) volatile and non-volatile metabolites inhibiting the growth of pathogenic microorganisms without requiring direct contact, and therefore the choice of experimental method solely depends on the experimental objectives. The dual culture method has received intensive use, as it can detect, in addition to antibiosis (whether estimated by the scale of Bell et al. [29] or by the action of volatile and non-volatile metabolites), the occurrence of a number of important phenomena, such as competence for the substrate and mycoparasitism. The cellophane method, by its very design, is only able to measure antibiosis through the action of non-volatile metabolites secreted into the culture medium.
Amplification of the internal transcribed spacer of rDNA and elongation factor
Analysis of PCR reactions for the amplification of an ITS fragment revealed the presence of 250 and 350 bp amplicons for the ITS1 and ITS2 primer pairs, respectively. These sizes coincide with those of the amplicons obtained by PCR from a reference strain of T. atroviride (IMI206040) (Figures 2 and 3).
Since the decade of 1990, the sequences of regions of ITS1 and ITS2 have been employed for assignment to the Trichoderma genus or the identification of its species [14, 19, 30-32]. These target sequences offer the advantage of highly sensitive and easy amplification, as they are present at copy numbers larger than 90 copies per genome.
O’Donnell et al. [33, 34], Buckler et al. [35] and Lieckfeldt and Seifert [36] have argued, however, that this marker is inadequate for this task due to the presence of ITS paralogues in other phytopathogenic or saprophytic fungal genera, although this phenomenon has not been detected in most species of Trichoderma/Hypocrea [37]. It has also been pointed out that ITS provide poor phylogenetic resolution for some groups, such as the Pachybasium B section.
Taking into account these objections, it was decided to also amplify a fragment of the EF1A translational elongation factor (Figure 4). These reactions yielded a 400 bp amplicon for both the test and reference strain samples. It should be stressed, however, that markers such as β-tubulin, ACT1 and EF1A itself do not provide optimal phylogenetic resolution across the entire genus or even the major genetic subtypes such as Pachybasium B, despite the fact that the study of the largest tef1 intron provided excellent resolution within the subtype and related taxa such as T. harzianum / H. subtype lixii species (Hypocrea lixii, T. harzianum, T. aggressivum, T. tomentosum Bissett, T. cerinum, T. velutinum, Hypocrea tawa) or the Hypocrea rufa (Pers.) P. group, which includes T. viride, T. atroviride and T. koningii. The data of Kindermann et al. [30] further support these results, evidencing that solely using a gene fragment is not sufficient for this purpose.
Taylor et al. [38] have proposed to base the concept of phylogenetic species on the agreement of five or more phylogenetic trees, a scheme hardly applicable to Trichoderma given the extreme variability of this taxon. Recently, studies combining the sequencing of ITS with the genes mentioned above have also included endochitinase genes in an attempt to increase the reliability of their conclusions [21, 39-42].
Despite the similarity of the results obtained in the present case for the amplification of ITS regions and the elongation factor between the test isolates and the reference strain of T. atroviridae (IMI206040), it was decided to sequence the obtained amplicons and perform a homology search against TrichoBLAST/GenBank® [24]. According to the resulting data, the sequence of these isolates is identical to those of T. asperellum. The observed similarity in amplicon size when compared to T. atroviridae probably stems, therefore, from the inclusion of both species into the H. rufa group, in agreement with the observations of Druzhinna and Kubicek [9] as well as Kindermann et al. [30].
Evaluation of the efficacy of Trichoderma spp. isolates
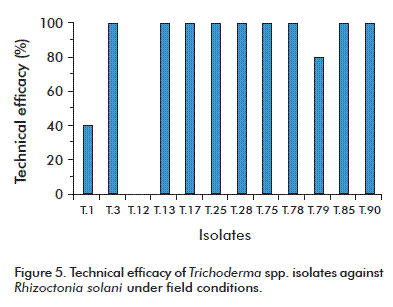
Under field conditions, 10 of the T. asperellum isolates exhibited a technical efficacy against R. solani higher than 80 %. Isolate T.1 exhibited a technical efficacy of 40 %, and isolate T.12 was not effective (Figure 5). These results highlight the importance of adequately screening isolates intended to be used as biological control agents.
The efficacy of Trichoderma spp. isolates as agents for the biological control of phytopathogens has been examined by many different authors. For instance, Veitia et al. [43] examined the feasibility of using Trichoderma spp. isolates for the biological control of R. solani in basil, obtaining an efficacy after 45 days of 60 to 90 % when growing the plants in trays under controlled conditions. On the other hand, Hoyos et al. [21] evaluated the efficacy of five isolates of T. asperellum against Sclerotium rolfsii Sacc. and R. solani under semicontrolled conditions, obtaining values higher than 90 % for isolates T.21 and T.71 and over 50 % for isolates T.109 and T.110. The results of the present study coincide with those of Martínez et al. [28], who obtained efficacy values higher than 80 % with isolates T.17, T.27, T.56, T.75 and T.78 of Trichoderma spp. against R. solani. Their results were especially impressive for isolates T.17 and T.27, which exhibited an efficacy higher than 90 % for plants grown in stainless steel trays. Similar results were published also by Reyes et al. [44], who obtained technical efficacies higher than 80 % for the control of R. solani with isolates T.78 and T.56 of T. asperellum, also using stainless steel trays.
Since isolates T.3, T.13, T.17, T.25, T.28, T.75, T.78, T.85 and T.90 were promissory, additional tests were run under open field and greenhouse conditions, in correspondence with Martínez et al. [28] as well as Reyes et al. [44]. This is relevant, even, for the selection of the method for mass production of these biopesticides.
In Cuba, Trichoderma spp. is used mainly for the control of soil-dwelling fungi. Strains C-66 and A-34 of T. harzianum have exhibited efficacies higher than 99 % against Rhizoctonia solani in bean (Phaseolus vulgaris L) [45]. In onion (Allium cepa L.), it was possible to decrease the incidence of R. solani through the application of T. viride [46]. However, not much research has been carried out in Cuba on the modes of action of this biological control agent, although acceptable in vitro antagonism has been obtained for T. harzianum against R. solani isolates from tomato (Solanum lycopersicum L.), pepper (Capsicum annuum L.) and carnation (Dianthus caryophyllus L.) [47]. It has also been shown that T. harzianum strains A-34 and C-66, as well as A-34 and A-53, exhibit mycoparasitic activity against R. solani isolated from bean [48] and rice (Oryza sativa L.) [49], respectively.
CONCLUSIONS
In the course of the present work, the use of molecular markers derived from ITS and the elongation factor as well as their sequencing has enabled the correct taxonomical assignment of a group of Trichoderma spp. isolates that can potentially be used for the control of R. solani isolates belonging to different anastomosis groups, including group AG-5, which is highly pathogenic for rice.
In addition, our data provide additional depth to existing knowledge about this genus, highlighting how the antifungal activity of Trichoderma spp. isolates can proceed through a variety of very specific modes of action, and offering perspectives for the application of said isolates and their metabolites. Specifically, the finding that these isolates antagonize their target phytopathogen through the secretion of non-volatile metabolites has very clear implications for their production by liquid fermentation.
Trichoderma spp. biopreparations, both in solid and liquid formulations, can preventively be applied in soils known to be infected with R. solani, taking advantage of the longer durability of the structures of the former (conidia, chlamydospores and mycelia) and their capacity to parasite Rhizoctonia sclerotia, which constitute the main structure for the propagation of this parasitic fungus. Our findings, therefore, can be used to significantly improve the process of selection, application and reproduction of Trichoderma spp. strains intended for biological control.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the support provided by a scholarship granted by the Public Health Secretariat (SEP) of Mexico, without which this work would not have come into fruition. We would also like to thank Dr. Alfredo Herrera Estrella, for making possible our stay as visiting scientists at the National Laboratory of Genomics for Biodiversity (Langebio) of the National Polytechnic Institute (IPN), under his direction. Likewise, we thank technicians María Isabel Blanco Campos and Noreidys Fernández Gálvez for their participation in the present study.
REFERENCES
1. Hjeljord L, Tronsmo A. Trichoderma and Gliocladium in biological control: An overview. In: Harman GE, Kubicek CP, editors. Trichoderma and Gliocladium. Vol. 2. Enzymes, Biological Control and Commercial Applications. London: Taylor and Francis Ltd.; 1998. p. 131-51.
2. Kullnig C, Mach RL, Lorito M, Kubicek CP. Enzyme diffusion from Trichoderma atroviride (= T. harzianum P1) to Rhizoctonia solani is a prerequisite for triggering of Trichoderma ech42 gene expression before mycoparasitic contact. Appl Environ Microbiol. 2000;66(5):2232-4.
3. Tondje PR, Roberts DP, Bon MC, Widmer T, Samuels GJ, Ismaiel A, et al. Isolation and identification of mycoparasitic isolates of Trichoderma asperellum with potential for suppression of black pod disease of cacao in Cameroon. Biol Control. 2007; 43(2):202-12.
4. Woo SL, Scala F, Ruocco M, Lorito M. The Molecular Biology of the Interactions Between Trichoderma spp., Phytopathogenic Fungi, and Plants. Phytopathology. 2006;96(2):181-5.
5. Nelson EE. Occurrence of Trichoderma in a Douglas-fir soil. Mycologia. 1982; 74(2):280-4.
6. Hoyos-Carvajal L, Orduz S, Bissett J. Genetic and metabolic biodiversity of Trichoderma from Colombia and adjacent neotropic regions. Fungal Genet Biol. 2009;46(9):615-31.
7. Cervantes MA. Microorganismos del suelo beneficiosos para los cultivos [Internet]. Madrid: Infoagro Systems, S.L.;2012 [cited 2012 Mar 11]. Available from: http://infoagro.com/hortalizas/microorganismos_beneficiosos_cultivos.htm.
8. Harman GE. Myths and Dogmas of Biocontrol Changes in Perceptions Derived from Research on Trichoderma harzianum T-22. Plant Dis. 2000;84(4):377-93.
9. Druzhinina I, Kubicek CP. Species concepts and biodiversity in Trichoderma and Hypocrea: from aggregate species to species clusters? J Zhejiang Univ Sci B. 2005;6(2):100-12.
10. Klein D, Eveleigh DE. Ecology of Trichoderma. In: Kubicek CP, Harman GE, editors. Trichoderma and Gliocladium. Vol. 1. Basic Biology, Taxonomy and Genetics. London: Taylor and Francis Ltd. 1998; p. 57-74.
11. Samuels GJ. Trichoderma: systematics, the sexual state, and ecology. Phytopathology. 2006;96(2):195-206.
12. Crop Protection Compendium (CPC)[Internet]. Wallingford: Centre for Agricultural Bioscience International (CABI); c2012 - [cited 2012 Mar 11]. Available from: http://www.cabi.org/cpc/.
13. Rifai MA. A revision of the genus Trichoderma. Mycology; 1969;(116):1-56.
14. Kuhls K, Lieckfeldt E, Samuels GJ, Meyer W, Kubicek CP, Börner T. Revision of Trichoderma section Longibrachiatum including related teleomorphs based on analysis of ribosomal DNA internal transcribed spacer sequences. Mycologia. 1997; 89(3):442-60.
15. Bissett J. A revision of the genus Trichoderma. I. Section Longibrachiatum sect. nov. Can J Bot. 1984;62(5):924-31.
16. Bissett J. A revision of the genus Trichoderma. II. Infrageneric classification. Can J Bot. 1991;69(11):2357-72.
17. Bissett J. A revision of the genus Trichoderma III. Section Pachybasium. Can J Bot. 1991;69(11):2373-417.
18. Samuels GJ. Trichoderma: a review of biology and systematics of the genus. Mycol Res. 1996;100(8):923-35.
19. Samuels GJ, Lieckfeldt E, Nirenberg HI. Trichoderma asperellum, a new species with warted conidia, and redescription of Trichoderma viride. Sydowia. 1999;51(1): 71-88.
20. Lieckfeldt E, Samuels GJ, Nirenberg HI, Petrini O. A morphological and molecular perspective of Trichoderma viride: is it one or two species? Appl Environ Microbiol. 1999;65(6):2418-28.
21. Hoyos-Carvajal L, Paola Chaparro, Miriam Abramsky, Chet I, Sergio O. Evaluation of Trichoderma spp. isolates against Rhizoctonia solani and Sclerotium rolfsii under in vitro and greenhouse conditions. Agron Colomb. 2008;26(3):451-8.
22. Dennis C, Webster J. Antagonistic properties of species-groups of Trichoderma: II. Production of volatile antibiotics. Trans Br Mycol Soc. 1971;57(1):41-8.
23. White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky J, White TJ, editors. PCR Protocols: A guide to Methods and Applications. San Diego: Academic Press; 1990. p. 315-22.
24. Kopchinskiy A, Komon M, Kubicek CP, Druzhinina IS. TrichoBLAST: a multilocus database for Trichoderma and Hypocrea identifications. Mycol Res. 2005;109(Pt 6):658-60.
25. Abbott WS. A method of computing the effectiveness of an insecticide. 1925. J Am Mosq Control Assoc. 1987;3(2):302-3.
26. Küçük Ç, Kivanç M. Mycoparasitism in the biological control of Gibberella zeae and Aspergillus ustus by Trichoderma harzianum. J Agric Technol. 2008;4(2):49-55.
27. Infante MD. Efecto antagónico de ocho aislamientos de Trichoderma frente a Rhizoctonia sp. in vitro [dissertation]. Tesis de diploma. La Habana: Universidad Agraria de La Habana; 2006.
28. Martínez B, Reyes Y, Infante D, González E, Baños H, Cruz A. Selección de aislamientos de Trichoderma spp. candidatos a biofungicidas para el control de Rhizoctonia sp. en arroz. Rev Protección Veg. 2008;23(2):118-25.
29. Bell DK, Wells DH, Markham CR. In vitro antagonism of Trichoderma species against six fungal plant pathogens. Phytopathology. 1982;72(4):379-82.
30. Kindermann J, El-Ayouti Y, Samuels GJ, Kubicek CP. Phylogeny of the genus trichoderma based on sequence analysis of the internal transcribed spacer region 1 of the rDNA cluster. Fungal Genet Biol. 1998;24(3):298-309.
31. Lieckfeldt E, Kullnig CM, Kubicek CP, Samuels GJ, Börner T. Trichoderma aureoviride: phylogenetic position and characterization. Mycol Res. 2001;105(3):313-22.
32. Dodd SL, Crowhurst RN, Rodrigo AG, Samuels GJ, Hill RA, Stewart A. Examination of Trichoderma phylogenies derived from ribosomal DNA sequence data. Mycol Res. 2000;104(1):23-34.
33. O’Donnell K. Ribosomal DNA internal transcribed spacers are highly divergent in the phytopathogenic ascomycete Fusarium sambucinum (Gibberella pulicaris). Curr Genet. 1992;22(3):213-20.
34. O’Donnell K, Cigelnik E, Nirenberg HI. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 1998;90(3):465-93.
35. Buckler IV ES, Ippolito A, Holtsford TP. The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics. 1997;145(3):821-32.
36. Lieckfeldt E, Seifert KA. An evaluation of the use of ITS sequences in the taxonomy of the Hypocreales. Stud Mycol. 2000;45:35-44.
37. Kullnig CM, Krupica T, Woo SL, Mach RL, Rey M, Benítez T, et al. Confusion Abounds Over Identities of Trichoderma Biocontrol Isolates. Mycol Res. 2001; 105(7):770-2.
38. Taylor J, Jacobson D, Fisher M. The evolution of asexual fungi: Reproduction, Speciation and Classification. Annu Rev Phytopathol. 1999;37:197-246.
39. Kullnig-Gradinger CM, Szakacs G, Kubicek CP. Phylogeny and evolution of the fungal genus Trichoderma: a multigene approach. Mycol Res. 2002;106(7):757-67.
40. Kubicek CP, Bissett J, Druzhinina I, Kullnig-Gradinger C, Szakacs G. Genetic and metabolic diversity of Trichoderma: a case study on South-East Asian isolates. Fungal Genet Biol. 2003;38(3):310-9.
41. Chaverri P, Samuels GJ. Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocreaceae): species with green ascospores. Stud Mycol. 2004;48:1-36.
42. Druzhinina IS, Kopchinskiy AG, Komon M, Bissett J, Szakacs G, Kubicek CP. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet Biol. 2005;42(10):813-28.
43. Veitia MM, García V, Izquierdo D, Porras A, Wong W. Control de Rhizoctonia sp. en Albahaca blanca (Ocimun basilicum L.) con Trichoderma harzianum cepa 34. Fitosanidad. 2000;4(1-2):67-70.
44. Reyes Y, Martínez B, Infante D. Evaluación de la actividad antagónica de trece aislamientos de Trichoderma spp. sobre Rhizoctonia sp. Rev Protección Veg. 2008;23(2):112-7.
45. González M. Utilización de Trichoderma spp. para el control de hongos patógenos de la semilla y del suelo en el cultivo del frijol [dissertation]. Tesis en opción al título de Máster en Protección Vegetal. La Habana: Universidad Agraria de La Habana; 2001.
46. Meléndrez J, Santana M, Herrera L. Estudio de la incidencia de Rhizoctonia solani Kühn en el cultivo de la cebolla (Allium cepa L.) en la zona de Banao. Centro Agrícola. 2002;29(2):51-4.
47. Sandoval I, López MO, Bonilla T, Tomas Y. Hongos del suelo que atacan al clavel y antagonismo in vitro con Trichoderma spp. Fitosanidad. 1998;2(3-4):41-4.
48. González M, Castellanos L, Ramos M, Pérez G. Efectividad de Trichoderma spp. para el control de hongos patógenos de la semilla y el suelo en el cultivo del frijol. Fitosanidad. 2005;9(1):37-41.
49. Alarcón I, Reyes T, Rodríguez G, Pupo AD. Efectividad in vitro de Trichoderma harzianum (Rifai) en el biocontrol de Rhizoctonia solani Kühn y Pyricularia grisea (Sacc.) en el cultivo del arroz (Oryza sativa L.). Fitosanidad. 2005; 9(3):57-60.
Received in March, 2012.
Accepted in July, 2012.
Danay Infante. Grupo de Fitopatología, Dirección de Protección de Plantas, Centro Nacional de Sanidad Agropecuaria, Censa. Autopista Nacional km 23½, AP 10, San José de las Lajas, Mayabeque, Cuba. E-mail: danay@censa.edu.cu.