Mi SciELO
Servicios Personalizados
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Biotecnología Aplicada
versión On-line ISSN 1027-2852
Biotecnol Apl vol.31 no.1 La Habana ene.-mar. 2014
REPORT
Isolation and cloning of the Phytolacca americana anti-viral protein PAP-I gene
Aislamiento y clonaje del gen de la proteína antiviral PAP-I de Phytolacca americana
Heba A Mahfouze1, Khalid A El-Dougdoug2, Badawi A Othman2, Mostafa A Gomaa1
1 Genetic Engineering and Biotechnology Division, Genetics and Cytology Department, National Research Center. Dokki, 12622, Egypt.
2 Virology Laboratory, Department of Agriculture Microbiology. Faculty of Agriculture, University of Ain Shams, Cairo, Egypt.
ABSTRACT
The pokeweed antiviral protein (PAP) isolated from Phytolacca americana and Phytolacca acinosa plants, inhibits protein translation by catalytically removing a specific adenine residue from the large rRNA of the 60S subunit of eukaryotic ribosomes. In this study, the P. americana PAP-I gene was isolated and sequenced, and further compared to other ribosome-inactivating proteins (RIPs) genes previously reported in GenBank®. Total DNA was extracted from the late summer leaves of P. americana. A polymerase chain reaction (PCR) 868 bp-long DNA product was obtained, using gene specific primers, based on the expected gene size. The eluted product was purified and cloned into the pTZ57R/T vector, and mobilized into the Escherichia coli strain DH5a. After sequencing, the analysis of the PAP-I PCR product showed 98 to 82 % nucleotide and amino acid 94 to 26 % homologies, respectively, compared to previously reported RIPs. A phylogenetic analysis confirmed that the amplified PAP-I gene corresponds to the single chain Type-I RIP (PAP-I).
Keywords: pokeweed antiviral protein, cloning, PCR, DNA sequencing, phylogenetic alignment.
RESUMEN
La proteína antiviral del ginseng (PAP), aislada de plantas de Phytolacca americana y Phytolacca acinosa, inhibe la traducción proteica mediante la remoción catalítica de un residuo de adenina específico, en la cadena mayor de la subunidad 60S del ARN ribosomal eucariótico. En este estudio se aisló y secuenció el gen PAP-I de P. americana, y posteriormente se comparó con los genes de otras proteínas inactivadoras de ribosomas (RIP), reportadas en GenBank®. Se extrajo el ADN total de las hojas tardías del verano de las plantas de P. americana y el fragmento de 868 pb correspondiente al ADN del gen se amplificó con el uso de cebadores específicos, mediante reacción en cadena de la polimerasa (PCR). El producto de la PCR eluido se purificó, se clonó en el vector pTZ57R/T, y se movilizó en células de Escherichia coli cepa DH5a. Tras la secuenciación del producto de la PCR del gen PAP-I, la secuencia mostró una homología nucleotídica de 98 a 82 % y aminoacídica de 94 a 26 %, con las RIP reportadas. El análisis filogenético confirmó que el gen amplificado corresponde a la RIP tipo I de simple cadena (PAP-I).
Palabras clave: proteína antiviral del ginseng, clonaje, reacción en cadena de la polimerasa, secuenciación de ADN, alineamiento filogenético.
INTRODUCTION
Many plants produce proteins that inactivate ribosomes by depurinating a highly conserved stem-loop structure in the 28S rRNA [1-4]. Single-chain ribosome-inactivating proteins (RIPs) such as PAP and the A chains of two-chain RIPs such as ricin remove an adenine base by specific cleavage of the N-glycosidic bond in the 28S rRNA. Ribosomes depurinated in this manner are unable to bind the EF-2/GTP complex and protein synthesis is blocked at the translocation step [5]. Three different kinds of RIPs, PAP-I, PAP-II, and PAP-S, have been previously purified from pokeweed (Phytolacca americana) plants. These proteins are similar in molecular mass (29, 30, and 29.5 kDa, respectively), but are expressed at different developmental stages and in different pokeweed tissues [6].
PAP protein has shown depurinating activity not only on pokeweed ribosomes, but also on other plants [7], mammalian, yeast, and bacterial ribosomes. In addition, PAP effectively inhibits infection by a number of different plant [8] and animal viruses, including the human immunodeficiency virus [9], and also inhibits growth of tumor cells [1, 10]. The exogenous application of small amounts of PAP to the surface of plant leaves completely prevents mechanical transmission of unrelated viruses to several different host plants.
PAP is stored in the cell wall matrix of leaf mesophyll cells and can readily be obtained from water extracts of macerated leaf tissue. In a previous study, a cDNA clone for PAP was isolated and introduced into potato plants by transformation with Agrobacterium tumefaciens, providing resistance against both mechanically- and aphid-transmitted potato viruses [11]. Recently, the nucleotidic and amino acid sequences of PAP-I, PAP-α, PAP-II and PAP-S were determined. An amino acid sequence comparison between PAP-I and PAP-S revealed 76 % homology [12]. It has been determined by N-terminus protein sequencing that PAP-II is less homologous. There are also evidence of isozyme forms of various PAP-S proteins displaying different properties, which would make one isoform more valuable than others for therapeutic applications, what motivated researchers to clone and sequence the cDNA of PAP-II protein [13].
In order to have a PAP-I clone able to be manipulated for research purposes, in the present study we isolated and cloned the full length cDNA of PAP-I from pokeweed leafs. Its homology with other reported antiviral proteins was also analyzed.
MATERIALS AND METHODS
Materials
P. americana and P. acinosa seeds were obtained from the Leibniz Institute of Plant Science and Crop Plant Research (IPK), Gatersleben, Germany. Seeds were sowed in 14 cm pots and kept in a greenhouse for approximately three weeks at 21-25 °C until germination.
Preparation of genomic DNA
Genomic DNA was extracted from spring leaves of P. americana and P. acinosa. Harvested leaves were frozen with liquid nitrogen and ground to a fine powder in a mortar with pestle. Two hundred microliters of a solution containing 0.5 M Tris-HCl (pH 8), 0.25 M EDTA and 5 % SDS was added to 100 mg of leaf powder, and the mixture was extracted with phenol saturated with 45 mM sodium citrate and 0.45 M NaCl. The isolated aqueous phase was extracted with phenol/chloroform (1:1), and then with chloroform/isoamyl alcohol (24:1) three times. The genomic DNA in aqueous phase was ethanol-precipitated with 0.3 M ammonium acetate, washed with 70 % ethanol, and dissolved in distilled water [14].
PCR amplification
Two specific sets of primers specific for full-length PAP-I gene were designed based on the sequences available in the GenBank® database (http://www.ncbi.nlm.nih.gov/genbank): A (Forward 5´-AAGAG AGTGGAAGGGCAGCCT-3´ and Reverse 5´-AGTACCCATTGGCACGGCCT-3´) and B (Forward 5-ATAAATACAATCACCTTCGATGTTG-3´ and Reverse 5´- AGAATCCTTCAAATAGAGTCACCAA-3´). The reaction mixture contained 50 ng of genomic DNA, 2.5 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCland Reverse 5´-AGTACCCATTGGCACGGCCT-3´) and B (Forward 5-ATAAATACAATCACCTTCGATGTTG-3´ and Reverse 5´- AGAATCCTTCAAATAGAGTCACCAA-3´). The reaction mixture contained 50 ng of genomic DNA, 2.5 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCland Reverse 5´-AGTACCCATTGGCACGGCCT-3´) and B (Forward 5-ATAAATACAATCACCTTCGATGTTG-3´ and Reverse 5´- AGAATCCTTCAAATAGAGTCACCAA-3´). The reaction mixture contained 50 ng of genomic DNA, 2.5 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl
Cloning of PAP-I DNA
The amplified products were analyzed on a 1 % agarose gel. Products of approximately 868 bp were eluted separately and reamplified. The resulting bands were further eluted, purified and cloned into linearized pTZ57R/T Easy vector. The Escherichia coli strain DH5α was transformed by pTZ57R/T Easy vector bearing a PAP-DNA insert. Transformants were grown at 37 °C in LB broth containing ampicillin (100 μg/mL) to a 0.4 optical density at 600 nm. Recombinants were isolated by alkali minipreparation from a group of viable transformants and the presence of the insert was confirmed by restriction digestion with BamH I and Sac I compared to control vector, and PCR amplification [15].
Sequencing and homology analyses
The full length nucleotide sequences of the PAP-I gene were aligned with other PAP proteins available at GenBank® database using the BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple alignments and phylogenetic analysis of PAP-I sequencing were carried out using the program CLC sequence viewer v. 6.8.1 (CLC bio, Denmark) and the DNAMAN v. 5.2.9 software (Madison, Wisconsin, USA), respectively. Homology trees were set up with the distance matrix using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) method [16].
RESULTS AND DISCUSSION
Cloning, sequencing and sequence alignment of the PAP-I gene
The PAP-I gene was isolated from P. americana and P. acinosa leaves using specific primer sets A and B. The fragments of PAP-I gene from both P. americana and P. acinosa plants, were 1188 and 868-bp long using primers A and B, respectively (Figure 1). These results were in an agreement with those of Honjo et al. [14]. The isolation of the DNA fragment (868 bp) from genomic DNA of P. americana leaves by PCR used two specific primers based on the known DNA sequence of the PAP-I gene [17], and subsequently, the PCR product of the PAP-I gene was obtained by using the specific primer set B.
The product of the expected size (approximately 868 bp) was first eluted from the agarose gel, purified and further amplified, and verified by agarose gel electrophoresis. An aliquot of the amplified product was cloned into the pTZ57R/T vector and mobilized into E. coli DH5α competent cells. White ampicillin colonies were selected for plasmid minipreparation using the plasmid minipreparation techniques. A recombinant plasmid bearing the correct insert of the PAP-I gene (868 bp) was validated using PCR and restriction analysis, followed by agarose electrophoresis. The fragment was shown to be excised from the plasmid backbone by BamH I and Sac I restriction. Similar results were reported by Chen et al. [18], when constructing by PCR two DNA fragments coding for PAP, with or without signal peptide, which were cloned into the pKK233-2 expression vector and transformed into E. coli JM109 cells. Similarly, Lodge and Kaniewski [11] used PCR to engineer convenient restriction sites (Bgl II at the 5´ end and Sma I at the 3´ end) into the full-length cDNA encoding PAP for inserting it into the plant transformation vectors pMON8443, carrying the PAP gene under the control of the enhanced 35S RNA promoter from the cauliflower mosaic virus (CaMV E35S), and pMON8442 and pMON8484 under the 35S RNA promoter from the figwort mosaic virus (FMV), respectively. Three other groups [19-21] extracted total RNA from the leaves of pokeweed (P. americana) plants which had been subcultured for 8 weeks for the same purpose. The first strand of cDNA was synthesized from the total RNA template with oligo (dT) 15 primers using MMLV reverse transcriptase. A 0.96 kb cDNA fragment was obtained after 30 PCR amplification cycles with two specific primers. The cDNA fragment was sequenced from two directions after cloning it into the pGEM-T Easy vector. The plant expression vector with the PAP cDNA was constructed for transferring the PAP cDNA into tobacco and rape to obtain transgenic plants resistant to a broad spectrum of viruses.
The resulting full length nucleotide sequence of the PAP-I DNA isolated from P. americana was submitted to GenBank® under the JX445170.1 accession number. Multiple alignment of PAP-I gene under study with seven sequences from GenBank® using the BLAST algorithm showed a 98 % homology with alpha-PAP (Accession No. D10600.1) [22], followed by 83 % with sequence X98079.1, while the lowest identity was 82 % for sequences with Accession Nos. AY572976.1, X55383.1, JX446580.1, AY547315.1 and AB071855.1 (Table 1). High homology (94-26 %) was shown in amino acid sequences to other RIPs. Multiple alignment of the PAP-I sequence obtained with antiviral proteins PAP-I, PAP-II, PAP-S and MAP, and statistical analysis of the DNA fragment PAP-I showed that PAP-I differ from other RIPs in base composition (A: 282, C:172, G:177 and T: 237), and nucleotides frequency (A: 0.325, C: 0.198, G: 0.204 and T: 0.273). The PAP-I gene contains on one open reading frame from nucleotide 57 to 812, coding for a 289 amino acids protein with an 33.761 kDa molecular weight and theoretical isoelectric point of 10.15. The amino acid composition data of PAP-I showed different compositions and frequencies of amino acids between isolates (Table 2 and Table 3). Twenty amino acids were detected on PAP-I starting with Alanine (A) and ending with Tyrosine (Y). Leucine (L) was found to be the major amino acid in PAP-I (35) for a 0.121 frequency, followed by Serine (S) (30; 0.104). Aspartic acid (D) was the less frequent aminoacid (4; 0.014). These results were comparable to those of Honjo et al. [14], who found that PAP genomic DNA sequence amplified from P. americana encoded a protein of 262 amino acids with a c-terminal 29 amino acids extrapeptide.
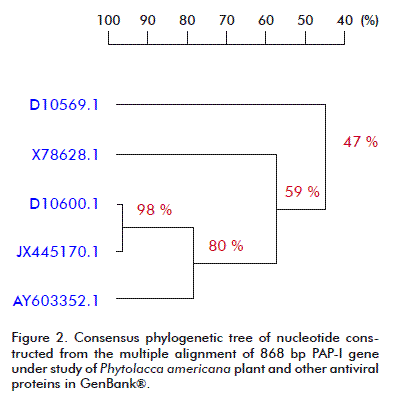
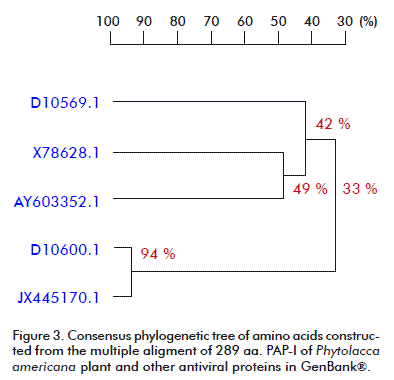
The phylogenetic analysis carried out for the nucleotide and amino acid sequences of PAP-I, and other RIPs available at the GenBank® database, by using the DNAMAN V 5.2.9 package (Madison, Wisconsin, USA) with the UPGMA method is presented in Figure 2 and Figure 3. A close relationship was found between our PAP-I isolate and D10600.1 (bootstrap value 98 % in nucleotide sequence and 94 % in amino acid homology) (Figure 2 and Figure 3). Poyet and Hoeveler [12] isolated and sequenced the PAP-I gene from P. americana, and characterized for the first time a com-plete cDNA encoding a pokeweed antiviral protein expressed in seeds. The PAP-S cDNA consisted of 1249 bp and codes for a mature protein of 262 amino acids. Its predicted amino acid sequence was more similar to PAP-I (76 %) than to PAP-II (31 %).
It is known from literature that PAP-S is more active in inhibiting protein synthesis than other members of the PAP family. For example, Zeng et al. [23] found that sequence similarities among these PAP-S, α-PAP and PAP-I with known three-dimensional structures are about 75-80 %. Wang et al. [24] isolated a 30 kDa PAP-II protein from leaves of P. americana, which inhibits translation by catalytically removing a specific adenine residue from the large rRNA of the 60S subunit of eukaryotic ribosomes. The protein sequence of PAP-II shows only 41 % identity to PAP and PAP-S, two other antiviral proteins isolated from pokeweed. Their isolated cDNA corresponding to PAP-II were introduced into tobacco plants, and transgenic tobacco plants correctly processed PAP II to its mature form as in pokeweed, and accumulated it to at least 10-fold higher levels than the wild-type PAP.
CONCLUSIONS
Many plants contain proteins that are capable of inactivating ribosomes and accordingly are called RIPs. These typical plant proteins currently receive a lot of attention in biological and biomedical research because of their unique biological activities toward plant, animal and human cells. In addition, evidence is accumulating that some RIPs play a role in plant defense and hence can be exploited in plant protection. The present study provided a characterized clone of the PAP gene sequence to be expressed for antiviral applications. Further research on the isolation of PAP full length DNA will be useful in constructing plant transformation vectors, to obtain transgenic plants resistant to relevant viral diseases.
REFERENCES
1. Stirpe F, Barbieri L, Battelli MG, Soris M, Lappi DA. Ribosome-inactivating proteins from plants: present status and future prospects. Biotechnology. 1992;10(4):405-12.
2. Salehzadeh A, Arasteh A. Expressing of rice ribosome inactivating protein as tool for treatment of cancer cells. Sci Res Essays. 2012;7(1):61-5.
3. Yang J, Jin GH, Wang R, Luo ZP, Yin QS, Jin LF, et al. Spinacia oleracea proteins with antiviral activity against Tobacco mosaic virus. Afr J Biotechnol. 2012; 11(26):6802-8.
4. Kataoka J, Miyano M, Habuka N, Masuta C, Koiwai A. A genomic gene for MAP, a ribosome-inactivating protein from Mirabilis jalapa, contains an intron. Nucleic Acids Res. 1993;21(4):1035.
5. Osborn RW, Hartley MR, Tumer NE. Dual effects of the ricin A chain on protein synthesis in rabbit reticulocyte lysate. Inhibition of initiation and translocation. Eur J Biochem. 1990;193(2):401-7.
6. Barbieri L, Aron GM, Irvin JD, Stirpe F. Purification and partia1 characterization of another form of the antiviral protein from seeds of Phytolacca americana L. (pokeweed). Biochem J. 1982;203(1):55-9.
7. Bonness MS, Ready MP, Irvin JD, Mabry TJ. Pokeweed antiviral protein inactivates pokeweed ribosomes; implications for the antiviral mechanism. Plant J. 1994;5(2):173-83.
8. Chen ZC, White RF, Antoniw JF, Lin Q. Effect of pokeweed antiviral protein (PAP) on the infection of plant viruses. Plant Pathol. 1991;40(4):612-20.
9. Zarling JM, Moran PA, Haffar O, Sias J, Richman DD, Spina CA, et al. Inhibition of HIV replication by pokeweed antiviral protein targeted CD4+ cell by monoclonal antibodies. Nature. 1990;347:92-95.
10. Smirnov S, Shulaev V, Tumer NE. Expression of pokeweed antiviral protein in transgenic plants induces virus resistance in grafted wild-type plants independently of salicylic acid accumulation and pathogenesis-related protein synthesis. Plant Physiol. 1997;114(3):1113-21.
11. Lodge JK, Kaniewski WK, Tumer NE. Broad spectrum virus resistance in transgenic plants expression pokeweed antiviral protein. Proc Natl Acad Sci USA. 1993;90(15):7089-93.
12. Poyet JL, Hoeveler A. cDNA cloning and expression of pokeweed antiviral protein from seeds in Escherichia coli and its inhibition of protein synthesis in vitro. FEBS lett. 1997;406(1-2):97-100.
13. Poyet JL, Radom J, Hoeveler A. Isolation and characterization of a cDNA clone encoding the pokeweed antiviral protein II from Phytolacca americana and its expression in E. coli. FEBS Lett. 1994;347(2-3):268-72.
14. Honjo E, Watanabe K, Tsukamoto T. Study on the expression of pokeweed antiviral protein in Escherichia coli as a fusion with maltose-binding protein. Bull Fac Agr Saga Univ. 1997;(82):83-90.
15. Sambrook E, Fritsch F, Maniatis T. Molecular cloning. A laboratory manual. 2nd Ed. New York: Cold spring Harbor Laboratory Press; 1989.
16. Sneath PHA, Sokal RR. Numerical taxonomy. San Francisco: W.H. Freeman and Company; 1973. p. 230-4.
17. Lin Q, Chen ZC, Antoniw JF, White RF. Isolation and characterization of a cDNA clone encoding the anti-viral protein from Phytolacca americana. Plant Mol Biol. 1991;17(4):609-14.
18. Chen ZC, Antoniw JF, Lin Q, White RF. Expression of pokeweed (Phytolacca americana) antiviral protein cDNA in Escherichia coli and its antiviral activity. Physiol Mol Plant Pathol. 1993;42(4):237-47.
19. Zhang H, Tian Y, Zhou Y, Dang B, Lan H, Song G, et al. Introduction of poke-weed antiviral protein cDNA into Bra-ssica napus and acquisition of transgenic plants resistant to viruses. Chin Sci Bull. 1999;44(8):701-04.
20. Chen GJ, Li S, Jian LJ, Bi CH, Guo ZP. Cloning of Pokeweed antiviral protein gene from Phytolacca acinosa and its transfer to pepper (Capsicum annuum L). Acta Hort Sin. 2008;35(6):847-52.
21. Cao B, Lei J, Chen G, Cao P, Liu X, Chen Q, et al. Testing of disease-resistance of pokeweed antiviral protein gene (PacPAP) in transgenic cucumber (Cucumis sativus). Afr J Biotechnol. 2011;10(36):6883-90.
22. Kataoka J, Habuka N, Masuta C, Miyano M, Koiwai A. Isolation and analysis of a genomic clone encoding a pokeweed antiviral protein. Plant Mol Biol.1992;20(5):879-86.
23. Zeng ZH, He XL, Li HM, Hu Z, Wang DC. Crystal structure of pokeweed antiviral protein with well-defined sugars from seeds at 1.8A resolution. J Struct Biol. 2003;141(2):171-8.
24. Wang P, Zoubenko O, Tumer NE. Reduced toxicity and broad spectrum resistance to viral and fungal infection in transgenic plants expressing pokeweed antiviral protein II. Plant Mol Biol. 1998;38(6):957-64.
Heba A Mahfouze. Genetic Engineering and Biotechnology Division, Genetics and Cytology Department, National Research Center. Dokki, 12622, Egypt. E-mail: hebaamn@yahoo.com.