Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista Cubana de Informática Médica
versión On-line ISSN 1684-1859
RCIM vol.9 no.2 Ciudad de la Habana jul.-dic. 2017
TRABAJOS ORIGINALES
Studying Protein kinases PKCζ and PKMζ with the Resonant Recognition Model. Implications for the study of Memory Mechanisms
Estudio de proteínas quinasas PKCζ y PKMζ mediante el Modelo de Reconocimiento Resonante. Implicaciones para el estudio de los Mecanismos de Memoria
Dr. Suria Valdés GarcíaI
DrC. José Luis Hernández-CáceresI
Lic.Damián Palmero ColmenaresI
I "Diez de Octubre," Medical Faculty, Havana Medical Sciences University, Havana, Cuba.
Corresponding author: cacerjlh@infomed.sld.cu
ABSTRACT
PKMζ is a brain-specific protein kinase that has been suggested as playing a key role in memory consolidation mechanisms. It is identical to catalytic portion of another protein kinase, PKCζ. Lacking the regulatory end, PKMζ is several times more active than PKCζ. However, knowledge about PKMζ mechanisms in memory consolidation is patchy, and sometimes contradictory. The resonant recognition model (RRM) might shed some light in understanding PKMζ role on memory consolidation. This is the first attempt in literature to apply the RRM to the study of PKMζ and PKCζ. We obtained that PKMζ presents a spectral peak at the resonant recognition frequency of fRRM= 0.063 (likely, corresponding to the infrared frequency of 3190 nm) and another peak at fRRM =0.211(950 nm in the near infrared). Peak at fRRM= 0.063 is also shared by PKCζ, and the peak at fRRM =0.211 is similar to the one recently reported in literature for regulatory proteins. We hypothesize that irradiating with a weak light infrared source at these frequencies would modify long term potentiation results. Finally, a scheme for resonant interactions in PKMζ and PKCζ is proposed.
Palabras claves: long term potentiation, protein kinases, resonant recognition model, bioinformatics.
RESUMEN
PKMζ es una proteína quinasa específica del cerebro que se ha sugerido que desempeña un papel clave en los mecanismos de consolidación de la memoria. Es idéntica a la porción catalítica de otra proteína quinasa, PKCζ. Al carecer de la porción regulatoria, PKMζ es varias veces más activa que PKCζ. Sin embargo, el conocimiento sobre los mecanismos de PKMζ in en la consolidación de la memoria es parcial, y a veces contradictorio. El modelo de reconocimiento resonante (RRM) podría esclarecer la comprensión del papel de PKMζ en la consolidación de la memoria. Este es el primer intento en la literatura para aplicar el MRR al estudio de PKMζ y PKCζ. Se obtuvo que PKMζ presenta un pico espectral a la frecuencia de reconocimiento resonante fRRM = 0,063 (probablemente, correspondiente a la frecuencia infrarroja de 3190 nm) y otro pico a fRRM = 0,211 (950 nm en el infrarrojo cercano). Pico en fRRM = 0,063 es también compartida por PKCζ, y el pico a fRRM = 0,211 es similar a la recientemente informado en la literatura para las proteínas reguladoras. Se plantea la hipótesis de que la irradiación con una fuente de luz infrarroja débil a estas frecuencias podría modificar los resultados de potenciación a largo plazo. Finalmente, se propone un esquema para interacciones resonantes en PKMζ y PKC.
Key words: potenciación a largo plaz, proteínas kinasas, modelo de reconocimiento resonante, bioinformática.
Introduction
Elucidation of memory mechanisms remains a central problemin Neuroscience. The discovery of long term potentiation-manifested as a substantial increase in synaptic efficacy after a tetanic stimulus- provided a model for studying memory mechanisms. Studies carried out by Eric Kandel and his grouprevealed that long term potentiation involves protein synthesis as well as structural changes in involved synapses.1
The possibility that a single molecule can be the main responsible for memory processes was suggested by Todd Sacktor, who found that PKMζ(a brain-specific protein kinase devoid of regulatory pseudo-substrate portion) is the responsible for triggering a cascade of events leading to sustained self-maintenance of up-regulated synaptic efficacy.2 Thus it has been suggested that inhibition of PKMζ in the hippocampus, insular cortex, and amygdala erasesseveral types of memories such as place memory, conditioned taste aversion, and fear memory, respectively, as well as counters long term potentiation. 3
Proposed scenarios for PKMζ mechanisms touch interaction of this protein with other proteins, as CREB-binding protein (CBP), the adaptor protein importin-α, the peptide ZIP, can be viewed under the light of protein interactions.4 A universal mechanism has been proposed for protein interactions that offers an alternative way to traditional views.
Thus Irena Cosic has developed the Resonant Recognition Model (RRM). According to this model, bio-photons are sent by proteins and can be recognized by substrates and receptors as well as by proteins participating in a common biological function. Proteins with a common biological function do share the same frequency, whereas interacting proteins share the same frequency and opposite phase.
We found no reports about RRM studies withPKMζ. We hypothesize that a RRM study on this protein could suggest new possibilities to appraise its role in memory consolidation.
Methods
Data:
In order to perform RRM analysis, a group of sequences were downloaded from UNIPROT at www.uniprot.org
PKMζ:
PKMζ is the C-terminal fragment of the brain-specific protein kinase PKCζ corresponding to amino acids positions from 184 to 592 (see Table 1). Lacking inhibition from the pseudo-substrate of the regulatory domain of PKCζ, PKMζ is a persistently active enzyme. Although PKM is usually thought of as a cleavage product of full-length PKC it has been reported that PKM is not formed in LTP by proteolysis but by gene expression of a brain-specific PKM mRNA, which is generated by an internal promoter within the PKC gene.5 Apparently Tetanic stimulation induces protein synthesis from the PKM mRNA, persistently increasing the levels of the kinase during LTP maintenance.
For resonant frequencies identification, several proteins sharing the same function were studied. Here we analyzed both PKCζand PKMζ sequences from the following twelve species:
1. U3IRA8 (U3IRA8_ANAPL) Anas platyrhynchos (Mallard) (Anas boschas) ánade real
2. U3K4C5 (U3K4C5_FICAL)Ficedula albicollis (Collared flycatcher) (Muscicapa albicollis) papamoscas collarino
3. E1BQN6 (E1BQN6_CHICK) Gallus gallus (Chicken) gallo/ gallina
4. G3RB70 (G3RB70_GORGO) Gorilla gorilla gorilla (Western lowland gorilla) gorila occidental de llanura o planicie
5. G3TKV6 (G3TKV6_LOXAF) Loxodonta africana (African elephant) elefante africano de sabana
6. G1MTR8 (G1MTR8_MELGA)Meleagris gallopavo (Common turkey) pavo salvaje
7. H0WJ10 (H0WJ10_OTOGA)Otolemur garnettii (Small-eared galago) (Garnett's greater bushbaby) gálago de Garnet
8. K7FJG4 (K7FJG4_PELSI) Pelodiscus sinensis (Chinese softshell turtle) (Trionyx sinensis) tortuga de caparazón blando de China
9. H0YXM1 (H0YXM1_TAEGU) Taeniopygia guttata (Zebra finch) pinzón cebra
10. Q05513 (KPCZ_HUMAN) Homo sapiens (Human) humano
11. Q02956 (KPCZ_MOUSE) Mus musculus (Mouse) ratón común
12. P09217 (KPCZ_RAT) Rattus norvegicus (Rat) rata parda
Each PKMC sequence in these twelve species contained exactly 592 amino acids. The PKMζ sequence was obtained from corresponding PKCζ by extraction of the fragment with amino acids from 184 to 592.
For comparison, the PKCZ segment from position 1 to 183 (containing the regulatory unit and the hinge) was also submitted to RRM analysis.
Resonant Recognition Model
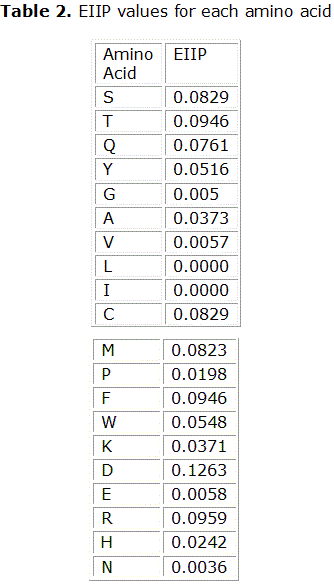
The RRM, proposed by Irena Cosic, postulates that interactions of proteins with receptors, peptide substrates and other proteinsare achieved through resonant energy transfer between involved molecules at the frequency specific for each observed function/interaction.6 This transfer of bio-photons at energies of the order of 10-20 J is the universal mechanism by which macromolecules and smaller peptides recognize each other.7 This vision is radically different from the idea of interactions of through van der Waals forces, hydrogen bonds and structural complementarity as the main role players in the molecular recognition by proteins. A key aspect of the RRM approach is to represent a protein’s primary structure as a numerical series. For this, each amino acid in the sequence is symbolized with the (numerical) value of a biologically relevant physical-chemical parameter. If the chosen parameter is suitable, it happens that proteins with the same biological function have a common frequency component in their Fourierspectra. This common frequency is considered to be a hallmark ofa protein’s biological function/interaction. After trying with different candidate parameters, it was found that the energy of delocalized electrons, calculated as the electron–ion interaction pseudo-potential (EIIP) of each amino acid residue is the best suited quantity for RRM analysis.8 EIIP values for each amino acid appear in Table 2. The EIIP parameter describes the average energy states of all valence electrons in a particular amino acid. Accordingly, the resulting numerical series represents the distribution of the free electron energies along protein’s backbone.
Once the numerical sequence is obtained, it is submitted to spectral analysis using the Fourier Transform (FT) to extract information pertinent to the biological function. In the frequency domain, the FT of an individual protein sequence will contain nonzero values for many frequencies. 9
However, if a cross spectral function is estimated for a group of proteins sharing one common frequency, the cross spectral function will have a nonzero value at this resonant frequency.
Since it can be expected that a given protein can display more than one function, it may happen that the cross spectrum of a group of orthologous proteins will exhibit more than one peak.
The multiple cross-spectral function obtained from a group of orthologous sequences with the same biological function has been named ‘consensus spectrum’. The presence of a distinct peak frequency in a consensus spectrum implies that this common frequency is related to the shared biological function provided the following criteria are met:
· One peak only exists for a group of protein sequences sharing the same biological function;
· No significant peak exists for biologically unrelated protein sequences;
· Peak frequencies are different for different biological functions.
Cosic has studied a large amount of proteins and concluded that each specific biological function of a given protein characterized by a single frequency. The RRM can be applied to the study of interactions of proteins with their targets (receptors, ligands and inhibitors) since it was found that interacting proteins and targets display the same characteristic frequency in their interactions. 6
Thus, the RRM characteristic frequencies represent a protein’s general functions as well as the mutual recognition between a particular protein and its target (receptor, ligand, etc.). As this recognition arises from the matching of periodicities within the distribution of energies of free electrons along the interacting proteins, it can be regarded as the resonant recognition.
It has been found that peptides attaching to proteins do share the same resonant frequency and exhibit opposite phase. Thus, abiding these two conditions is regarded as a hallmark for protein-protein interaction.
The primary amino acid sequences were transformed into a numerical series following the Resonant Recognition Model (RRM) methodology. For it, to each of the 20 amino acids in the entire sequence an electron-ion interaction potential (EIIP) value was assigned (Table 2).
The obtained numerical series was treated as a time series. Power spectrum was estimated for each sequence using a SciLab program based on Fourier analysis. For finding the consensus spectrum, all the twelve spectral vectors were submitted to scalar cross multiplication. The obtained product is considered as the consensus spectrum.
The RRM frequency was converted to a true electromagnetic frequency by determining the appropriate wavelength using the empirical function proposed by Cosic: 6 fRRM = 201/λ.
Results
PKCζ. As apparent from Figure 1, a clear single peak was found at the frequency of fRRM= 0.063. Unlike many other protein sequences, this protein kinase exhibits only one prominent peak. Likely, this would correspond to the infrared frequency of 3190 nm.
PKMζ. We obtained, that PKMζ shares some features of PKCζ whereas differing in others (Figure 2).As observed, PKMζ shows a prominent peak at the same frequency of fRRM =0.063. However, when amplitudes were compared, the peak seems to be more than 100 times higher. At the same time, a smaller second peak appears at fRRM =0.211(950 nm in the near infrared).
Regulatory+Hingedomains
As it can be seen from figure 3, the regulatory domain with the hinge exhibit a peak at fRRM =0.33, corresponding to 609 nm (yellow light).
May regulatory unit be a "pseudo-substrate"?
A consensus spectrum was obtained from combining 12 PKMζ and 12 sequences of the regulatory + hinge domain. As it can be noticed from figure 4 there is a peak at fRRM= 0.063. This support the idea that PKMζ can combine with the regulatory domain even after PKCζ cleavage.
Discussion
So far, RRM remains as a Hypothesis,10 accepted by a small fraction of research community. However, it brings plausible predictions to a group of experimental data, such as a nice correspondence between spectral density changes and results of mutagenesis experiments, excellent correlation between theoretical fRRMand absorption frequency for light-absorbing proteins, biological function of different synthetic peptides designed via RRM, as well as bio-photon emissions of proteins in solution in full accordance with the Cosic model, among others.11
If proven true, Cosic´s model will herald a "true revolution in bioinformatics", as suggested by Mae.
It is worth of notice that RRM analysis considers only primary structure of the protein backbone, which makes it radically different from most approaches in structural biology and bioinformatics.
Our results can be summarized as follows:
- There is a resonant frequency at fRRM= 0.063 that is shared by both PKCζ and PKMζ. This frequency is not prominent at regulatory+ hinge domains. The peak corresponding to PKMζ is more than 100 times higher.
- A smaller, but significant peak appears at fRRM=0.22. This peak is not observable in PKCζ nor in the regulatory + hinge domains.
- The regulatory + hinge domains may interact with PKMζ at the resonant frequency of fRRM= 0.063.
These results do agree with the idea of PKCζ and PKMζ sharing a common function, supported in this case by the common frequency at fRRM= 0.063. This frequency is also used for recognition between PKMζ and the regulatory unit.
It is recognized that PKMζ is much more active than PKCζ, and the higher amplitude of the peak could be a corroboration of this in RRM terms. However, the common view for this difference is the idea that the regulatory domain act as a pseudo-substrate for the catalytic domain. However, in RRM analysis only the primary sequence is taken into account and this is enough for suggesting marked differences in activity levels.
On the other hand, interacting proteins do share the same frequency, only with different phases. The fact that the peak at fRRM= 0.063 is common between regulatory+hinge domains and PKMζ suggests that the two portions of the cleaved PKCζ would interact leading to a reduced function. This could explain why PKMζ is obtained in vivo by direct synthesis and not from the cleavage of PKCζ.
Our results also suggest that the peak at fRRM=0.212 (950 nm in the near infrared), is related to the function of PKMζ as a signal protein. Dotta et al. suggested that "According to Cosic’stheory to predict macromolecular bioactivity, the 950-nm band is associated with molecules involved with signaling activities within the cell as well as cell proliferation. These processes are activated by forskolin and inhibited by PD8059." 7 Thus this peak probably reflects the condition of PKMζ as a signal protein, a function that is shared with PKA, and enhanced with forskolin.
Similarly, it is to be clarified which function is the peak at fRRM =0.33 -observed for the regulatory and hinge domain- is associated to. In figure 5 we are comparing the classical view on PKCζ andPKMζ with the RRM view based on results from this work.
PKCζ can be viewed as a resonance system emitting/receiving infrared light 3190 nm. PKMζ is much more active at this main wavelength, and also is tuned to near infrared at 913 nm. The regulatory and hinge domains are tuned to yellow light (609 nm). At the same time, they do interact with PKMζ through infrared light 3190 nm.
Further studies would involve the RRM study of proteins putatively interacting with PKMζ, in order to elucidate if the condition of common frequency and opposite phase is met.
Finally, RRM suggest two ways of interacting with PKCζ. One is via the design of peptides12 and the other via weak electromagnetic radiation at the above-proposed wavelengths. The effect of them upon LTP could suggest new aspects of PKCζ in memory mechanisms.)
On the light of present results it is expected that monochromatic infrared light 3190 nm should modify LTP. At this stage is difficult to predict whether the effect will be inhibiting or enhancing. Apparently the experiment would be technically affordable, either using infrared LED or Laser sources.
Conclusion
La toma de decisiones se presenta muchas veces en los procesos administrativos, especialmente en condiciones de incertidumbre, en condiciones de riesgo y en condiciones de conflicto. En esta última la toma de decisiones resulta mucho más compleja, en el sentido que la decisión adoptada no depende únicamente del tomador de decisiones, sino además de la influencia de su oponente. Muchas son las aplicaciones de la Teoría de Juegos, entre las que destaca los juegos contra la naturaleza, ejemplificado en este trabajo a través de un problema práctico, en el que se decide invertir un presupuesto en recursos informáticos en la Facultad de Tecnología de la Salud de la Universidad de Ciencias Médicas de Santiago de Cuba para ser explotados durante el quinquenio 2016 – 2020, y para lo cual, independientemente de la plataforma informática establecida, se puedan adaptar y reutilizar al máximo.
Dicho problema fue modelado como un problema de PL, y resuelto con el apoyo del Solver, una herramienta para la solución de este tipo de problemas de optimización, incorporada en el Microsoft Excel.
Se pudo hallar una solución óptima, en el que se recomendaba invertir solamente en hardware para estaciones de trabajo, y en hardware y componentes de red, lo que garantizaría por lo menos un 72.08% de adaptación de los recursos invertidos, cualquiera que sea la plataforma informática establecida en la Facultad.
References
1. Kandel ER, Dudai Y, Mayford MR.The molecular and systems biology of memory. Cell, 2014; 157:163-186.
2. Sacktor TC. How does PKMzeta maintain long-term memory? Nature reviews. Neuroscience, 2011; 12: 9-15.
3. Von Kraus LM, Sacktor TC, Francis JT. Erasing sensorimotor memories via PKMzeta inhibition. PLoS One 5 2010: 11-15.
4. Ko HG, Kim JI, Sim SE, Kim T, Yoo J, Choi SL, Baek SH, Yu WJ, Yoon JB, Sacktor TC, Kaang BK. The role of nuclear PKM. in memory maintenance. Neurobiology of Learning and Memory (2016);169, doi: http://dx.doi.org/10.1016/j.nlm.2016.06.010
5. Serrano P, Yao Y, and Sacktor TC. Persistent Phosphorylation by Protein Kinase M Maintains. Late-Phase Long-Term Potentiation.The Journal of Neuroscience, February 23, 2005; 25(8):1979 –1984.
6. Cosic I. Macromolecular Bioactivity: Is It Resonant Interaction BetweenMacromolecules?-Theory and Applications IEEE. Transactions on Biomedical Engineering. DECEMBER 1994; 41(12): 1101-1114.
7. Blake T, Dotta J, Murugan M, Karbowski RM, Persinger MA. Shifting wavelengths of ultraweak photon emissions from dying melanoma cells: their chemical enhancement and blocking are predicted by Cosic’s theory of resonant recognition model for macromolecules Naturwissenschaften 2014; 101:87–94. DOI 10.1007/s00114-013-1133-3.
8. Lazovic J. Selection of amino acid parameters for Fouriertransform-based analysis of proteins. CABIOS COMMUNICATION 1996; 12 (6): 553-562.
9. Cosic I, Lazar K, Cosic D. Prediction of Tubulin resonant frequencies using the Resonant Recognition Model (RRM). IEEE Trans on NanoBioscience. 2015; 12:491–6. doi:10.1109/TNB.2014.2365851. ).
10. Cosic I. The Resonant Recognition Model of Bio-molecular Interactions: possibility of electromagnetic resonance. Polish Journal of Medical Physics and Engineering 2001; 7 (1): 73-87.
11. Murugan NJ, Karbowski LM, Persinger MA.Cosic D. Resonance Recognition Model for Protein Sequences and Photon Emission Differentiates Lethal and Non-Lethal Ebola Strains: Implications for Treatment. OpenJBiophysics.2014; 5:35.
12. Cosic I, Pirogova E.Bioactive Peptide Design using the Resonant Recognition Model. Nonlinear Biomedical Physics, 2007; 1(7), doc: 10.1186/1753-4631-1-7. ).
Recibido: 20 de julio de 2017.
Aprobado: 5 de septiembre de 2017.