INTRODUCCIÓN
For the purpose of exploring the metabolic potentials of Mediterranean Sea snails, the pigmented bacteria associated with the sea snail Thais sp., collected from Bahary shore were isolated using three different culture media: MZM, MZM of 25% strength and OLIGO medium. Plates were incubated at 37 °C and 25 °C separately, and the grown colonies were observed and collected daily during three weeks. A red-pigmented isolate was selected for the study which was identified, based on phenotypic traits (growth on MZM, MacConkey agar, Triple sugar iron (TSI) medium, Gram reaction, electron microscopy, and using VITEK®2 system) as well as 16S rRNA gene sequence analysis, as Vibrio sp. with 98% phylogenetic similarity toV. rhizosphaeraestrain MSSRF3, V. mangrovistrain MSSRF38, andV. ruberstrain VR1, the isolate was designated as V. sp. SHF which was subsequently proven to have both pks-1 and nrps genes.
Since the majority of bacterial genes coding for polyketide synthases (PKSs), non-ribosomal peptide synthetases (NRPSs), and hybrids (PKS-NRPS) remain silent in the absence of particular stimuli; hence, to study the effect of some stimuli on mRNA expression of pks-1 andnrps genes responsible for the biosynthesis of prodigiosin and other bioactive compounds, RT-qPCR was adopted to investigate the effect of sublethal concentrations of some stress-iducing chemicals (SDS 0.1% (vol/vol), H2O2 0.01%(vol/vol); DMSO 1% (vol/vol); imipenem 0.05 μg/mL; ofloxacin 0.05 μg/mL; and Chromium VI oxide, copper oxide, lead oxide, and nickel III oxide were also tested at a concentration of 0.001 μg/mL) as well as the co-culture (membrane filtered culture supernatant of Staphylococcus aureusATCC 6538P 0.5% (vol/vol)) on the up-regulation and down-regulation of both genes. gyrB was used as a reference gene for normalization. Considerable up-regulation ofnrpsexpression was observed (in terms of 2-ΔΔCt) for copper oxide treated cultures ofV.sp. SHF (39-fold) compared with the control cultures, followed by DMSO (22-fold), chromium VI oxide (19-fold), SDS (17-fold), and imipenem antibiotic (10-fold). Down-regulation ofnrps, at the mRNA level, was observed for ofloxacin treated cultures. Forpks-1 gene expression, interestingly maximum up-regulation was observed in cultures treated with DMSO (14-fold) followed by imipenem treated cultures (8.9-fold). Also, a twofold increase was observed with ofloxacin and copper oxide-treated cultures. Down-regulation was observed when cultures were treated with SDS, lead oxide, or cell-free culture supernatant ofS. aureusATCC 6538P.
For the analytical characterization of the red pigment of V. sp. SHF, the dried extract of the pigment was dissolved in methanol before being analyzed using UV-vis spectral analysis and LC-MS spectrometry. The UV-vis spectral analysis showed a maximum absorbance at 470 nm under alkaline conditions and at 525 nm under acid conditions. The LC-MS analysis showed a fraction separated at a retention time of 29.6 min with a mass to charge ratio (m/z) of 322.8 according to the negative ionization scanning mode and a m/z of 322.9 according to the positive ionization scanning mode. Such m/z ratios almost coincide with the m/z of 323 of prodigiosin; hence, the pigment was identified as prodigiosin- like pigment. To our knowledge, this is the first report on the isolation of aVibriostrain from a gastropod able to synthesize prodigiosin.
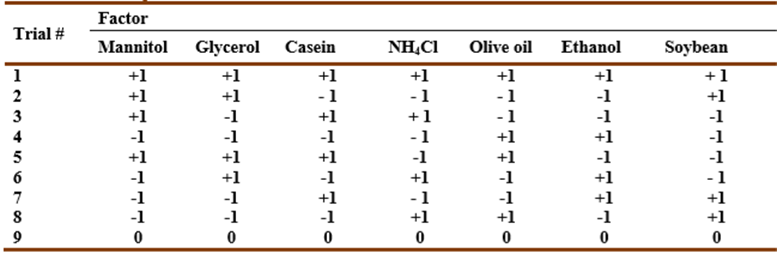
The effecs of several compounds on the pigment production was evaluated. For this purpose, prodigiosin concentration was calculated using absorbance at 490 nm and the extinction coefficient 51.3×103 L/(g cm). Supplementing MZM medium (pH 7) with mannitol as a carbon source enhanced growth and pigment production (7 mg/L) by an almost twofold increase compared to control cells after two days at 30 °C. Casein caused a 1.5-fold increase in pigment production. Replacing yeast extract with casein or tryptone resulted in a 1.7 and 1.4-fold increase, respectively. However, replacing peptone with tryptone reduced pigment production by 60%. Palm oil followed by L-cysteine, glucose, ammonium chloride, and starch addition decreased pigment production. Plackett-Burman fractional factorial design was applied to study the effect of seven variables (mannitol, glycerol, casein, ammonium chloride, olive oil, ethanol, and soybean meal) in nine combinations on pigment production in MZM (pH 7). The pigment production was optimized to 7 mg/L after two days in MZM cultures containing 0.5% mannitol and 2.5% soybean meal, and statically incubated at 30 °C.
Prodigiosin belongs to the family of red-pigmented prodiginines considered as bioactive secondary metabolites characterized by a common pyrrolyl dipyrromethene skeleton containing a common 4-methoxy, 2-2 bipyrrole ring system. Bacterial prodiginines are classified into linear and cyclic derivatives. The red pigment prodigiosin was isolated fromVibrio psychroerythreus, Vibrio gazogenes, andVibrio ruber. Other producers of this compound includePseudomonas magneslorubra, Alteromonas rubra, Hahella chejuensis, and differentSerratiaandStreptomycesspecies (Khanafariet al., 2006). Prodigiosin possesses bioactivities as antimicrobial, antitumor, and antimalarial agents (Kleinet al., 2017). Moreover, it was reported as an immunosuppressant, sunscreen, and a safe natural food dye. Bacterial prodigininss and their synthetic derivatives are effective proapoptotic agents against various cancer cell lines including multi-drug resistant cells with little or no toxicity towards normal cell lines. This includes several hematopoietic cancer cell lines, cervical carcinoma cell lines, as well as colorectal, lung, gastric, ovary, lymphoma, neuroblastoma, and breast cancer cell lines (Guryanovet al., 2020). Several prodigiosin derivatives with lower toxicity like GX15-070 have been clinically used. Due to the low fermentation yield and high production cost, its price was up to $500/mg (Xuet al., 2011).
The genusVibriocomprises a diverse group of heterotrophic marine bacteria of whichV. gazogenes,V. ruber, andVibrio rhizosphaerae(Rameshkumar & Nair, 2009) are the onlyVibriospecies reported to produce red pigments. From culture-dependent studies, vibrios appear in tissues and organs of various marine algae and animals, for example, abalones, bivalves, corals, fish, shrimp, sponges, squid, and zooplankton (Thompsonet al., 2004). Up to our knowledge, no study in literature have shown an association of vibrios with gastropods in general and the genusThaisin particular. Therefore, more studies should be directed to explore the microbiome of Mediterranean invertebrates, specially pigmented bacteria, and their metabolic potentials.
Thaisor the rock shell belongs to classGastropda, subclassNeogastropoda, familyMuricidae, subfamilyThaidinae(Claremontet al., 2013). Only a few studies have been conducted to explore its potentiality to serve the field of marine natural products or even other fields of applied sciences. The antimicrobial activity ofThais tissotiandThais haemastomaextracts was reported byKumaranet al. (2011)andSmaldoneet al. (2014). However, no previous report has dealt with isolation of microorganisms producers of bioactive compound from the microbiome of this gastropod.
Real-time quantitative polymerase chain reaction qPCR technology allows quantification and genotyping of pathogens, methylated DNA and microRNA analysis, validation of microarray data, allelic discrimination and genotyping (detection of mutations, analysis of single nucleotide polymorphism and microsatellites, identification of chromosomal alterations) and validation of drug therapy efficacy (Prada-Arismendy & Castellanos, 2011). Reverse transcription quantitative PCR (RT-qPCR) can also provide semi-quantitative results relative to controls (without standards) and hence enhancing gene expression studies (Kralik & Ricci, 2017). Unluckily, most of the identified bioactive compound gene clusters are silent under standard laboratory growth conditions. Effective methods for eliciting the production of new bioactive compounds include genetic engineering, mutagenesis, the one strain many compounds approach, or treatment with epigenetic modifiers (Netzkeret al., 2015). Hence, RT-qPCR can be used prominently in studying the effect of different stimuli on the expression of genes responsible for bioactive compounds production, and detecting gene expression differences as small as 23% between samples with lower coefficients of variation. It is even proven to be 1000-fold more sensitive than dot-blot hybridization and can detect even a single RNA copy (Wong & Medrano, 2005). The main aim of the current work was to optimize the prodigiosin-like pigment production by the study strain V. sp. SHF and to check the effect of sublethal concentration of some stress inducing chemicals on the mRNA expression of pks-1 andnrpsresponsible for the biosynthesis of prodigiosin and other bioactive compounds, using RT-qPCR.
MATERIALS AND METHODS
Isolation of bacteria associated with Thais snail
Snail samples were collected in February, 2016 from Bahary Beach, Alexandria, in sterile double plastic bags, then refrigerated at 2-4 °C. For bacterial isolation, the external surface was swabbed, and the flesh part was homogenized in 5 mL sterile seawater before applying the pour plate method described byFeby & Nair, (2010). Three different isolation media were used: MZM medium (a modification of Zobell marine medium) of the following composition (g/100 mL): Peptone 0.5, Yeast extract 0.1, and agar-agar 1.5. To enhance the isolation of oligotrophic bacteria, MZM of 25% strength and OLIGO medium (Feby & Nair, 2010) containing (g/100 mL): tryptone 0.005, yeast extract 0.0005, sodium glycerophospahte 0.001, and agar-agar 1.2 were used. Medium constituents were dissolved in seawater and pH was adjusted to 7 ± 0.2 using 1 N HCl and 1 N NaOH solutions. Plates were incubated at 37 °C and 25 °C separately, and the grown colonies were observed and collected daily during three weeks.
Identification of Vibrio sp. SHF isolated from Thais sp.
Phenotypic traits were determined according to growth on MZM, MacConkey agar, Triple sugar iron (TSI) medium slants, and using VITEK®2 system (Biomerieux, France) which was adopted for most of the biochemical investigation.
For electron microscopic examination, the cell pellet, obtained by the centrifugation of 3 mL of overnight MZM culture of V. sp. SHF, was mixed with 1mL of 4F1G fixative solution at 4 °C for 3 hr after which the specimen was post-fixed in OsO4 (2% in the same buffer) at 4 °C for 2 hr after which samples were washed in the buffer and dehydrated through a graded series of ethanol at 4 °C. Subsequently, the specimen was dried by means of a critical point method, mounted using carbon paste on an AL- stub and coated with gold up to a thickness of 400Ǻ in a sputter - coating unit (JFC-1100 E) (Pejman et al., 2015). Morphology observations were performed in Jeol JSM- 5300 scanning electron microscope operated between 15 and 20 keV.
For genomic DNA extraction, a single colony was picked from an MZM plate, inoculated into 5 mL MZM broth, and incubated overnight at 30 °C. 1 mL of each culture was centrifuged at 4500 xg for 20 min and cell pellets were collected. DNA was extracted using i-genomic BYF DNA Extraction Mini Kit. The integrity of the isolated DNA was confirmed by gel electrophoresis (using Power PacTM gel electrophoresis system (BIORAD, U.S.A). Chromosomal DNA was subjected to polymerase chain amplification reaction (PCR) using 2x MY TAQTM RED MIX. Primers were designed to amplify a 1500 bp fragment of the 16s rDNA region (Tamuraet al., 2007). The forward primer (F1) sequence was 5’-AGAGTTTGATCCTGGCTCAG-3’ and the reverse primer (R1) sequence was 5’-GGTTACCTTGTTACGACTT-3’. The PCR mixture consisted of 10 μM of each primer (1 µL), 2 μg of chromosomal DNA (2 µL), 8.5 µL ddH2O, and 12.5 µL of 2x My Taq Red mix. The PCR was carried out for 35 cycles (with a predenaturation step at 95 °C for 5 min), with the steps: 95 °C for 30 s, 50 °C for 30 s and 72 °C for 1 min. A final extension at 72 °C for 10 min was included. Amplification products were analyzed by agarose gel electrophoresis.
DNA sequence analysis was obtained using an ABI PRISM 377 DNA Automated Sequencer and ABI PRISM BigDye Terminator Cycle Sequencing (Perkin-Elmer, U.S.A). The PCR product was sequenced using the PCR primer pairs used for amplification. Sequences of 16S rRNA genes, for comparison, were obtained from the NCBI database (May, 2017). BLAST program was used to assess the DNA similarities and perform multiple sequence alignments. The molecular phylogeny was analyzed through multiple sequence alignments using ClustalX2 software. The phylogenetic tree was reconstructed by SeaView software.
Detection of pks-1 and nrps genes in V.sp. SHF
The purity and concentration of extracted DNA (eluted in 50 μL of the elution buffer) was checked by measuring the nucleic acid A260/A280 ratio using NanoDrop spectrophotometer also used to determine the concentration. Subsequent PCR conditions were optimized according to a concentration of 400 ng/mL of DNA. PCR amplification to detect the presence of polyketide synthase type I and non-ribosomal peptide syntethases (NRPS) genes (pks-1 andnrps, repectively) was carried out as described byAyuso-Sacido & Genilloud (2005). The following degenerate oligonucleotides were used: (a) K1F: 5’-TSAAGTCSAACATCGGBCA-3’, M6R: 5’-CGCAGGTTSCSGTACCAGTA-3’ to detectpks-1 gene (1200-1400 bp); (b) A3: 5’-GCSTACSYSATSTACACSTCSGG-3’, A7R: 5’-SASGTCVCCSGTSCGGTAS-3’ to detectnrpsgene (700-800 bp). PCR mixtures were prepared as mentioned before. Amplifications were then performed according to the following profile: 5 min at 95 °C and 35 cycles of 30 s at 95 °C, 30 s at 44.5 °C for K1F/M6R, 1 min at 45.9 °C for A3F/A7R, and 1 min at 72 °C, followed by 10 min at 72 °C. PCR amplification was confirmed by agarose gel electrophoresis (Tamuraet al., 2007).
Growth conditions
For growth and prodigiosin production, MZM (pH7) was prepared as sterile 2 mL aliquots in 24-well plates, inoculated with 2% inoculum of previously prepared seed culture (OD655 ~ 1), then incubated at 30 °C for two days, cells were then separated by centrifugation for 20 min at 4500 xg.
2.5. Studying the effect of some stress inducing chemocals on the expression of pks-2 and nrps in V. sp. SHF using RT-QPCR
The effect of some stress-inducing chemicals on gene expression was evaluated at sublethal concentrations. Cultures were treated by adding SDS 0.1% (vol/vol), H2O2 0.01%(vol/vol), DMSO 1% (vol/vol), imipenem 0.05 μg/mL, and ofloxacin 0.05 μg/mL to MZM broth. Chromium VI oxide, copper oxide, lead oxide, and nickel III oxide were also tested at a concentration of 0.001 μg/mL. According toFilkinset al. (2015), co-cultivation of two microbes can also enhance gene expression; hence, 100 μL of a membrane filtered (0.22 μm pore size) culture supernatant of Staphylococcus aureusATCC 6538P was tested.
Treated cell pellets were collected for total RNA extraction using easy-REDTM total RNA extraction TRIzol kit. Subsequently, extracted RNA was reverse transcribed into cDNA using Power cDNA synthesis kit, before applying RT-qPCR using REALMODTMGH Green Real-Time PCR master mix. Gene gyrB was used as a housekeeping gene for normalization. Sequences for the forward and reverse primers forgyrB gene were: 5´-GACGATGATCCGGTGGTAGC-3´ and 5´-CGATGATACCATCTTCGAGAC-3´, respectively. At the end of the reaction, the absence of non-specific amplification was checked using melting curves before the analysis. Trials were triplicated and duplicated forpks-1 and nrpsgene targeting specimens, respectively.
For data analysis, gene expression up-regulation or down-regulation was reported in terms of 2-ΔΔCt (Raoet al., 2013). Ct is the threshold cycle value that reflects the number of amplification cycles at which fluorescence generated within the reaction was significantly detected above background fluorescence. ΔCt was calculated by subtracting the threshold cycle value Ct of the housekeeping gene from Ct of the target gene. ΔΔCt was calculated as follows:
ΔΔCT= ΔCT (a target sample) - ΔCT (a reference sample).
The result of this method is presented as the fold change of target gene expression in a target sample relative to a reference sample normalized to a reference gene. The relative gene expression is usually set to one for reference samples because ΔΔCt is equal to zero and therefore 20 is equal to one.
ΔCt values were statistically analyzed using IBM SPSS 18 software. The non-parametric tests were adopted due to the small sample size; hence, Kruskal-Wallis H test was applied to assess data significance. Then a series of Mann-Whitney-U tests were used to conduct pairwise comparisons between the control group and other groups separately. The significance of the results was judged at the 5% level (Nahm, 2016).
Analytical characterization of V.sp. SHF red pigment
The pigment was extracted from cell pellets and purified as described by Ibrahim et al. (2014). The dry residues were redissolved in 2 mL of methanol, then subjected to ultraviolet-visible (UV-vis) spectral analysis and liquid chromatography-mass spectrosmetry (LC-MS).
UV-vis spectral analysis
UV-vis spectral analysis was conducted in the wavelength range from 400-600 nm using HeλiOS β spectrophotometer (Unicam, England) at 400-600 nm to ensure purity (Andreyeva & Ogorodnikova, 2015).
Liquid chromatography-Mass spectrometry (LC-MS)
Methanol extract of the red pigment was analyzed by electrospray ionization mass spectrometry (ESI-MS) using Agilent technologies 6420 Quad LC/MS system (Germany). The extracted pigment was analyzed using scanning LC (liquid chromatography) targeting fractions of molecular weight ranging from 100 to 1000 Da. Eclipse Plus C18 column (150×4.5 mm, 3.5 μm) was used with a 0-100% linear gradient in 36 min (A: 0.01 m ammonium acetate, pH 7, B: 100% acetonitrile). Five microliters were injected for both positive and negative ionization modes separately. The flow rate was 0.2 mL/min. Separation conditions were as follows: at zero time, mobile phase ratio (vol/vol) was 90% acidified H2O2 (0.1% formic acid), and 10% acetonitrile, finally after 36 min mobile phase ratio was 100% acetonitrile.
Table 1 Plackett-Burman matrix design representing seven independent variables. zero represents the original concentration, -1 represents the low concentration level, and +1 represents the high concentration level for each component.
Determination of prodigiosin-like pigment concentration
The bacterial growth in each trial was measured spectrophotometrically at 655 nm using a 96-well microtiter plate reader (BIORAD, U.S.A). Ethanolic extraction of the red pigment was conducted as described byRakhet al.(2017). Prodigiosin concentration was calculated using absorbance at 490 nm and the extinction coefficient 51.3×103 L/(g cm) (Andreyeva & Ogorodnikova, 2015). The resultant molar concentration was expressed as µg/mL.
RESULTS AND DISCUSSION
Isolation and Identification of the study isolate
After five days of incubation, a dark red pigmented colony developed on OLIGO agar plates, then it was picked and checked for purity (Fig. 1a). The bacteria grew faster on 25% strength MZM that was consequently selected for further studies (Fig. 1b). Cells were single swollen non-sporing motile Gram-negative rods (0.9 µm in length and 0.4 µm in width (Fig. 1c). Growth and color intensity increased at pH 5.Biochemical characterization was maily based on VITEK® 2 system using ID-GNB card, growth on MZM, growth on TSI slants, Gram staining, and KOH test (Table 3).
Table 3 Some morphological and biochemical characteristics of isolate AK-103
| Test | Result |
| Morphology | |
| Colony morphology | red small, rounded colonies with irregular margins after three days of growth on MZM, |
| Cell morphology | thick rods, with rounded ends, arranged as single cells |
| Motility | + |
| swarming | - |
| Gram staining | - |
| KOH test on MZM - | - |
| KOH test on MaCconkey's agar - | + |
| Growth conditions | |
| Growth at 4°C | - |
| Growth at 30°C | ++ |
| Growth at 37°C | + |
| Growth at 50°C | weak |
| Growth at pH 5-9 | + |
| Growth at 0% NaCl (Müller-himton agar and nutrient broth) | -/weak |
| Growth at 3% NaCl | + |
| Growth at 6 and 12% NaCl | - |
| Growth on TSI facultative anaerobe | |
| Butt color yellow | |
| Slant color red | |
| Gas production + | |
| Glucose fermentation + | |
| Sucrose - | |
| lactose - | |
| H2S production - | |
| Sugar fermentation | |
| Adonitol* | - |
| l-Arabitol* | - |
| D-Glucose* | - |
| D-Maltose* | - |
| Lactose (48hr on MacConkey's agar dissolved in sea water) | +/- |
| D-Mannitol* | - |
| D-Mannose* | - |
| D-sorbitol* | - |
| Sucrose* | - |
| Trehalose* | - |
| Tagatose* | - |
| Palatinose* | - |
| D-Cellobiose* | - |
| Hydrolytic activity | |
| Cellulose | - |
| Casein | + |
| Starch | - |
| Sodium dodecyl sulfate | + |
| Tri-calcium phosphate | + |
| Blood haemolysis (24hr at 37°C) | + (complete hemolysis) |
| Other biochemical tests | |
| Catalase | + |
| Citrate utilization* | - |
| H2S production* | - |
| Oxidase | - |
| Pellicle formation in liquid static cultures of MZM | + |
| Urease* | - |
| Phosphatase* | - |
| L-lactate alkalinization* | - |
| Succinate alkalinization* | - |
| L-malate assimilation* | - |
| L-lactate assimilation* | - |
| Malonate utilization* | - |
| 5-Keto-D-gluconate fermentation* | - |
| L-histidine assimilation* | - |
| Lysine decarboxylase* | - |
| Decarboxylase base* | - |
| Ornithine decarboxylase* | - |
| Glycine arylamidase* | - |
| Tyrosine arylamidase* | - |
| L-proline arylamidase* | - |
| L-pyrrolydonylarylamidase* | - |
| Ala-Phe-Pro arylamidase* | - |
| Glutamyl arylamidase pNA* | - |
| Glu-Gly-Arg arylamidase | - |
| Alpha-galactosidase* | - |
| Alpha-glucosidase* | - |
| Beta-alanine arylamidase pNA* | - |
| Beta-xylosidase* | - |
| Beta-galactosidase* | - |
| Beta-N-acetyl galactosaminidase* | - |
| Beta-N-acetyl glucosaminidase* | - |
| Beta-glucoronidase* | - |
| Beta-glucosidase* | - |
| Gamma-glutamyl transferase* | - |
| Courmarate* | - |
| Ellman* | - |
| O/129 Resistance* | - |
*: VITEK ® 2 system; +/-: weak pink color; 0/129: 2,4-diamino-6,7-di-isopropylpteridine phosphate

Fig. 1 Growth and morphological characteristics of the bacterium isolated in the present study: (a) MZM agar plates, (b) MZM broth, (c) Scaning electron micrograph of cells.
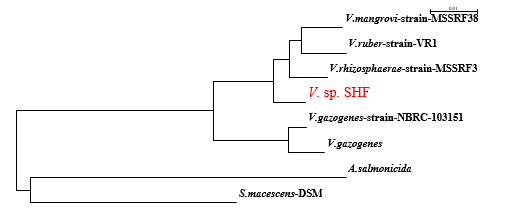
In comparison with the 16S rRNA gene, sequences held in GenBank indicated that the isolate was phylogenetically related to members of genus Vibrio with 98% similarity to V. rhizosphaerae strain MSSRF3, V. mangrovi strain MSSRF38, and V. ruber strain VR1 (Fig. 2). The strain was designated as V. sp. SHF. The 16S rDNA nucleotide sequence of V. sp. SHF was submitted to the NCBI GenBank, and was given the GenBank accession numbers: MK074974. Up to our knowledge, this is the first report for the isolation of a Vibrio species from the marine Thais sp.
Effect of some stimuli on expression of pks-1 and nrps in V. sp. SHF using RT-qPCR
Analysis of PCR amplification products shows thatV. sp. SHF has bothpks-1 andnrps genes. Fragments of 700-800 bp and 1200-1400 bp were obtained fornrpsand pks-1 genes, respectively (Fig. 3b and c). These two genes are reported to be involved not only in prodigiosin production but are also responsible for the synthesis of polyketide and non-ribosomal peptide bioactive compounds in general (Gomeset al., 2016).

Fig. 3 Gel electrophoresis of PCR amplified nrps (lane b) and pks-1 (lane c) from V. sp. SHF isolated from Thais sp. DNA ladder (lane a)
In bacteria, the majority of genes coding for polyketide synthases (PKSs), non-ribosomal peptide synthetases (NRPSs), and hybrids (PKS-NRPS) remain silent in the absence of particular stimuli (Brakhageet al., 2008). Hence, in this study, pks-1 andnrpsmRNA expression was evaluated in response to some stimuli.gyrB was used as a reference gene for normalization. In the current study, more than a 4-fold change in gene expression level was considered significant (Raoet al., 2013). The addition of stress-inducing compounds such as heavy metals, H2O2, antibiotics, or DMSO was expected to increase the biosynthesis of certain secondary metabolites. Furthermore, co-cultivation with cell-free culture supernatant ofS. aureusATCC 6538P, was also evaluated. Considerable up-regulation ofnrpsexpression was observed (in terms of 2-ΔΔCt) for copper oxide treated cultures ofV.sp. SHF (39-fold) compared with the control cultures, followed by DMSO (22-fold), chromium VI oxide (19-fold), SDS (17-fold), and imipenem antibiotic (10-fold) (Fig. 4). Imipenem was chosen as one of the antibiotics to whichV.sp.SHF was sensitive. Down-regulation ofnrps, at the mRNA level, was observed for ofloxacin treated cultures.
Forpks-1 gene expression, interestingly maximum up-regulation was observed in cultures treated with DMSO (14-fold) followed by imipenem treated cultures (8.9-fold) (Fig. 4). Also, a twofold increase was observed with ofloxacin and copper oxide-treated cultures. Down-regulation was observed when cultures were treated with SDS, lead oxide, or cell-free culture supernatant ofS. aureusATCC 6538P.
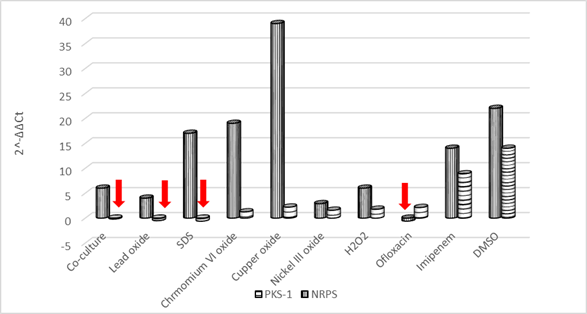
Fig. 4 Effect of some stimuli on pks-1 and nrps expression using RT-qPCR: Red arrows point out the down-regulation state.
The induction ofpks-1 andnrpsexpression by imipenem comes in agreement with the finding ofSeyedsayamdost (2014)who showed that sublethal concentrations of trimethoprim caused a global activation of secondary metabolism by inducing at least five biosynthetic gene clusters (mal, bhc, tha,andhmq), including new metabolites whose structures have not been determined. Similarly, other β-lactam, cephalosporin, fluoroquinolone, or DNA-alkylating antibiotic families at low concentrations seem to have transcriptional effects rather than growth inhibition. The report ofChenet al. (2000)framed the enhancing effect of DMSO on microbial secondary metabolites production, an observation that may explain the 22-fold increase inpks-1 gene expression and the 16-fold increase innrpsgene expression reported in the current study in DMSO treated cultures.
Table 4 Descriptive statistics of the responses of control and treated groups

N * : total number of trials.
The statistical significance of the effect of the studied stimuli on the level of mRNA expression ofpks-1 gene (as checked by IBM SPSS 18 statistics software using Kruskal-Wallis H test at 0.05 level of significance) was calculated as 0.02 (less than 0.05), while no significance was recorded for the same stimuli affecting thenrpsgene groups (Table 5).
Table 5 Stimuli affecting significantly the level of pks-1 and nrps gene expression as assessed by Kruskal-Wallis H test

The significance of the effect of each treatment on the level of mRNA expression ofpks-1 andnrpsgenes was further checked separately using a series of Mann-Whitney U tests to conduct pairwise comparisons between the control groups and each of the other treated groups. The Addition of DMSO, ofloxacin, imipenem, andS. aureuscell-free supernatant showed marginal significance (P = 0.05) on the level ofpks-1 gene expression while none of the studied stimuli showed a statistically significant effect onnrpsgene expression (Table 6). As reported byNahm (2016), the smaller is the sample size, the less is the statistical power of non-parametric tests. Hence the inability of the adopted tests to detect a significantp-value for the regulation ofnrpsgene expression does not mean an actual absence of significance.
Table 6 Significance of the effect of the different stimuli on the level of pks-1 and nrps gene expression as assessed by Mann-Whitney U test pairwise comparison of the control group and each treated group
Analytical characterization of V. sp. SHF red pigment
UV-vis spectral analysis
The spectroscopic analyses of the red pigment ofV. sp. SHF with UV-vis spectral analysis, under acidic conditions, showed the pigment was red with symmetrical broadband with a slight right-sided shoulder, and a peak at 525 nm. Under alkaline conditions, the pigment solution was orange-yellow, with a symmetrical band with a peak at 470 nm. At neutral conditions, the spectral peak was at 426 nm. Such pH-dependent color change is characteristic of prodigiosin produced bySerratia marcescensaccording toAndreyeva & Ogorodnikova (2015). The spectral analysis of the red pigment seemed to be close to those reported byIbrahimet al. (2014)who reported 535 nm for the acidic prodigiosin (produced byS.marcescenssolution and 465 nm for the alkaline one. Also,Bharmalet al. (2012)recorded a sharp spectral peak at 534 nm under acidic conditions. Similarly, maximum absorbance of the pigment at the pH values 2, 7, and 9 was found to be 535 nm, 458 nm, and 469 nm, respectively, as reported byFaraaget al. (2017).
Liquid chromatography-Mass spectrometry (LC-MS)
LC-MS analysis ofV. sp. SHF red pigment showed a fraction separated at a retention time of 29.6 min with a mass to charge ratio (m/z) of 322.8 according to the negative ionization scanning mode and a m/z of 322.9 according to the positive ionization scanning mode (Fig.5). Such m/z ratios almost coincide with that of prodigiosin produced byV. gazogenes(324) (Gummadidalaet al., 2016), andS. marcescens(323.5) (Kumar & Aparna, 2014). Similarly,Sumathiet al. (2014)reported a m/z of 324 for S.marcescensNPLR1 prodigiosin. Consequently, the red pigment was identified as a prodigiosin-like pigment.
According toSilvaet al. (2012), the substance produced byS. marcescens,which showed maximum UV absorbance at 534 nm and a m/z of 323, was characterized as prodigiosin. Moreover,Yanget al. (2013)described a red pigment, with a m/z of 323, produced byMicrocystis aeruginosa, as being prodigiosin.
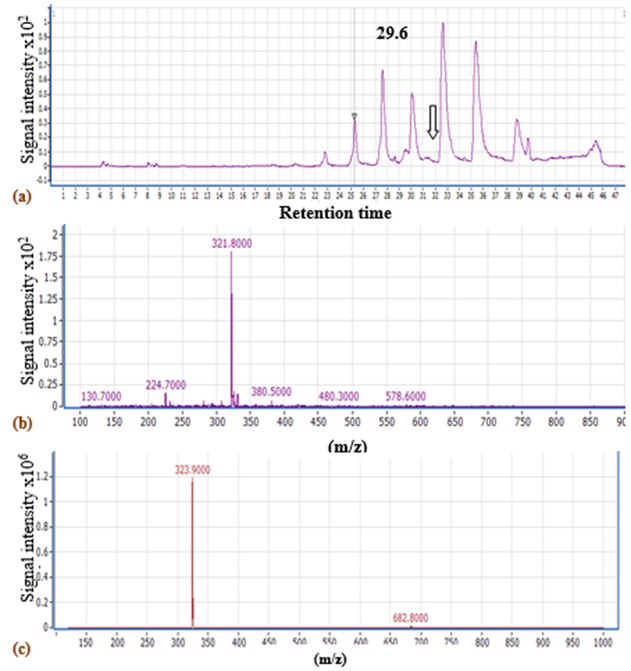
Fig.5. LC-MSchromatogram of V. sp. SHF DMSO extract using (a) negative ionization scanning mode (100-1000 Da) (The arrow refers to the fraction separated at 29.6 min), LC-MS chart of the peak showing the expected m/z of prodigiosin, using (b) negative inonization mode and (c) positive ionization mode.
Production of prodigiosin-like pigment by V. sp. SHF in response to different compounds
Supplementing MZM medium with mannitol (5 mg/L) as a carbon source enhanced growth and pigment production (7 mg/L) by an almost twofold increase compared to control cells (Fig. 6). Similar data were noticed byKurbanogluet al.(2015). Casein caused a 1.5-fold increase in pigment production. Replacing yeast extract with casein or tryptone resulted in a 1.7 and 1.4-fold increase, respectively. However, replacing peptone with tryptone reduced pigment production by 60%. The reduced pigment production in presence of glucose comes in agreement with the previous studies ofPhatake & Dharmadhikari, (2016)as well asBharmalet al. (2012), who explained such reduction in prodigiosin production byS. marcescensbased on lowering the pH by glucose, or catabolic repression.
Palm oil followed by L-cysteine, glucose, ammonium chloride, and starch addition decreased pigment production (Fig. 6). For glucose and starch, the decreased pigment production was associated with higher bacterial growth.Bharmalet al. (2012)showed that no prodigiosin was detected after 24 hr incubation when palm oil or starch was added to the medium, however, higher production was obtained after 72 hr only for the cultures supplemented with palm oil. Also in the same study higher production was attained after the addition of mannitol in agreement with the current study. However,Bharmalet al. (2012)reported higher prodigiosin production byS. marcescensupon the addition of 0.1% ammonium chloride which reduced the production in the current study.
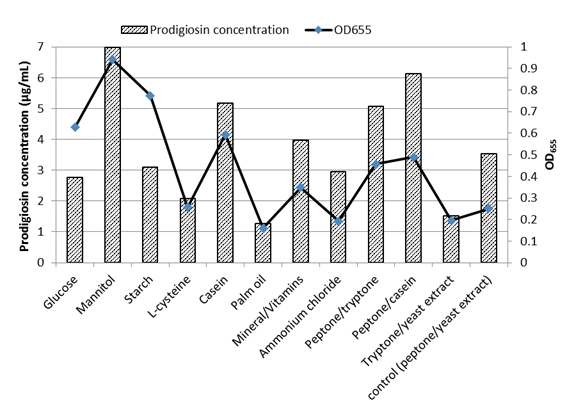
Fig.6 Effect of supplements added to MZM medium on prodigiosin-like pigment production and growth of V. sp. SHF
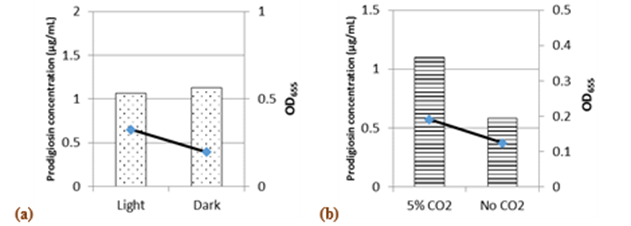
Incubation in the dark or the presence of white light did not affect pigment synthesis, although growth was better in light than in dark (Fig.7a). However,Phatake & Dharmadhikari, (2016)showed that the maximum level of pigment by S.marcescenswas achieved with white light incubation. In the presence of 5% CO2, 1.88 and 1.5-fold increases were recorded in pigment production and growth, respectively (Fig.7b).
Optimization of the prodigiosin-like pigment synthesis by V.sp. SHF using Plackett-Burman fractional factorial design
A total of seven variables were checked regarding their effects on pigment production (Table 7).
Table 7 Plackett-Burman design matrix representing the coded values for seven independent variables and their responses; -1 represents absence and +1 represents the concentration added.
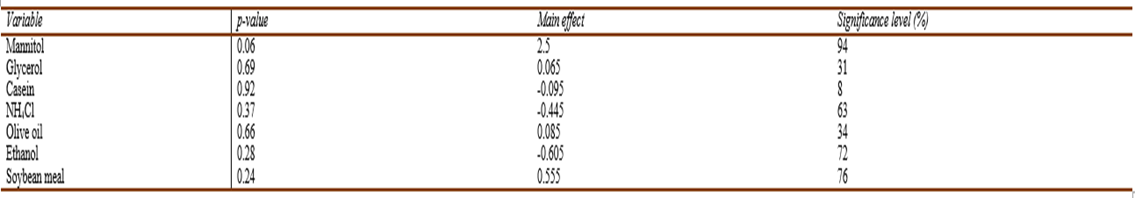
The Main effects plot of variables (Fig.8) demonstrated a significant positive effect of mannitol and soybean meal. Regression analysis of the experimental data was accomplished using Microsoft Excel (2013). The model coefficient of determination R = 0.99 with R-Squared (R2) = 0.98 and adjusted R2 = 0.94 implies that our model could explain 98% of the total variation, which in turn indicates a satisfactory representation of the process by the model (Elkenawyet al., 2017). According to Castro et al. (1992), since Plakett-Burman is a screening design; hence, 70% Confidence level is a useful guidepost for detecting significant effects. Thep-value was used to assess the significance of the studied factors. The results proved that mannitol, showed a confidence level of 94% followed by soybean meal which had a less significant effect as indicated by the confidence level of 76% (Table 8).
According toElkenawyet al. (2017), maximum production byS. marcescens(870 unit/cell) was achieved at 22 °C and pH 9 with the addition of 1% (wt/vol) peptone to 1% (vol/vol) crude glycerol resulting from biodiesel industry after six days of incubation.Canget al. (2000)reported ammonium chloride to affect negatively prodigiosin production byS. marcescens, as for our study. In the current study, olive oil addition seemed insignificant, however,Wei & Chen, (2005)reported an enhanced increase in prodigiosin yields (from 152 mgL-1 to 578 mgL-1) when the modified Luria Bertani broth was supplemented with 4% olive oil.

Fig.8 Main effect plot describing the significant variables affecting pigment production by V. sp. SHF using Plackett-Burman experimental design.
Table. 8 The main effect, significance level (%), and p-value for the determination of significant variable for pigment production by V. sp. SHF using Plackett-Burman experimental design
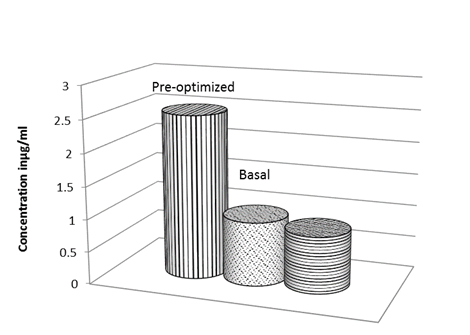
Based on these results obtained from Plackett-Burman experiment, MZM supplemented with 0.5% (wt/vol) mannitol and 2.5% (wt/vol) soybean meal was predicted to be near optimum for pigment production. A validation experiment was conducted, in which pigment concentration was estimated to conduct quantitatively a comparison among the pre-optimized, anti-optimized and basal media. The pre-optimized medium pigment yield was 2.5-fold higher than that of the basal medium and 3-fold higher than that of the anti-optimized medium (Fig. 9).
CONCLUSION
Considering the several reports discussing microbial prodigiosin production, mostly bySerratia,as well as prodigiosin production optimization due to its immense value as antimicrobial, antitumor, and antimalarial agent; and also as an immunosuppressant, sunscreen, and a safe natural food dye; however, this report can be considered as one of a few reports dealing with prodigiosin production by indigenous marineVibrioisolate, highlighting the necessity to explore bacteria associated with Mediterranean invertebrates in general and snails in particular for their ability to produce biologically active compounds of medicinal and industrial applications. The current study recommends adopting several integrated analytical techniques, including infra-red, nuclear magnetic resonance, and tandem mass spectrometry, to check the possibility that the isolated compound is structurally a new member of a known family of bioactive compounds. The enhanced prodigiosin production achieved through Plackett-Burman statistical design by supplementing the production medium with mannitol and soybean meal implies that utilization of industrial wastes like soybean meal is a vital tool to improve the performance of the bacterial system which helps to increase the yield of its products economically and also solves an environmental disposal problem. Moreover, herein, it was shown that the silent treasures of biosynthetic genes includingpks-1 andnrpscan be up-regulated through the addition of sublethal concentrations of stress-inducing agents like antibiotics, DMSO, heavy metals, or through co-cultures; hence, more varied stress-inducing strategies should be tried with this respect.