Introduction
With the recent advent of DNA probe technology, (a number of) many selective oligonucleotide probes that interact with specific DNA sequences in biological species have been identified, and used to provide a new type of selective biorecognition element.1
Label-free electrochemical DNA sensors mainly rely on the redox-active properties of DNA bases. Guanine and adenine are oxidized at much lower positive potentials than thymine and cytosine whose oxidation potentials are near the potential of oxygen evolution.2
Thus, the most active redox nitrogenous bases in DNA are guanine and adenine, which are more easily detected than thymine and cytosine. The oxidation of guanine and adenine, become the biomarkers of DNA sensors. Sequence-specific hybridization events have been monitored based on the oxidation signal of guanine.3
Common probe immobilization schemes include attachment of biotin-functionalized probes 4, self-assembly of organizing monolayers of thiol functionalized probes onto gold 5-7 and transducers carbodiimide covalent binding to an activated surface.3,8,9
The self-assembly of terminally thiol-labelled oligonucleotides offers a simple and elegant method to modify gold surfaces. This approach provides a unique probe structure flexibility that allows nearly a 100% hybridization efficiency.6 The modification the gold surface with organic thiols compound opens numerous opportunities for the construction of modified electrodes by using an Au-S interaction.
On the other hand, carbon electrodes are particularly attractive for sensing applications because of they are low cost, wide working potential window, good electrical conductivity, and relatively low background current for the development of electrochemical platforms.6 Graphite composite electrodes can also be easily modified, allowing the incorporation of different components such as ligands, enzymes, cofactors, mediators, and catalysts. Therefore, there is particular interest in using these electrodes in the development of sensors for the detection of biologically relevant molecules such as virus DNA.
Label-free electrochemical detection schemes greatly simplify the sensing protocols, as they eliminate the use of indicators. Moreover, the assay safety is improved, since the indicators are usually toxic, or carcinogenic compounds.
The first indicator-free scheme was introduced by Wang et al.2,6 The hybridization was detected by monitoring the decrease of the guanine peak of the immobilized probe. The use of label-free techniques has the added advantages of reducing costs and avoiding the need for sample pre-treatment.
The gag gene H1Gag1584 primer is a G-rich oligonucleotide (AAA GAT GGA TAA TCC TGG G) with 6 guanines, GC content 42% and guanine residue at 3´position.
The gag gene H1Gag1584 is a primer, it was used to that recognize a fragment of the gag gene in the heteroduplex mobility assay (HMA) sub-typing kit (NIH AIDS Research and Reference Reagent Program, USA) for study genetic variability for the HIV-1 env, gag and pol structural genes in Cuba 10 and sub-Saharan African cities.11 The primer H1Gag1584 (59-AAA GAT GGA TAA TCC TGG G-39) was used for nested PCR.
The aim of this work was to be able to test the usefulness of the biotin-modified 19-base oligonucleotide probe (biotinylated gag H1Gag1584) as a molecular recognition element of proviral DNA HIV-1 detection in two, free labels electrochemical detection systems.
Materials and methods
Reagents
Nuclease-free water, streptavidin (essentially salt-free, lyophilized powder, ≥13 units/mg protein), EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide, ≥98,0 %), and NHS (N-hydroxysuccinimide, >98,9 %) were purchased from Sigma-Aldrich (USA).
The probe used to monitor the impedimetric response of the electrode was equimolar solution of 1 mmolL-1, K3[Fe(CN)6]/K4[Fe(CN)6] in Phosphate Buffer at pH 6,9, 20 mmolL-1, and KCl 0,1 mol L-1, were purchased from Sigma-Aldrich (USA).
Solution of biotin and oligonucleotide were prepared in phosphate buffer at pH 6,9, 20 mmolL-1. Oligonucleotide for the biotinylated H1Gag1584 gag gene (sequence 5'AAA GAT GGA TAA TCC TGG G 3', sequence for the gag gene, identical to those of the ELI reference strain of the Los Alamos gene bank. Strain ELI, Genbank accession number K03454), and complementary oligonucleotide (c-DNA, sequence 5'CCC AGG ATT ATC CAT CTT T 3'), synthesized by (Exxtend Biotechnology Ltda) (oligonucleotide-Biotin, Brazil). The oligonucleotide employed encodes, a region of the HIV-1 H1Gag1584 gag gene, whose sequence is AAA GAT TAA TCC TGG G. This is one of the most conserved regions, which varies little between groups of HIV-1.12
Complementary DNA solution of concentration 2,5 10-12 molL-1 was prepared in buffer 0,1 molL-1 NaCl, Tris-Cl pH8, 0,1 molL-1 EDTA.13
Equipment
The voltammetric method was developed using a PalmSens instrument (Palm Instruments B.V., The Netherlands) operated in square-wave mode and coupled to a computer running PalmSens PC software. The graphite-epoxy working electrode modified with graphene oxide (EGIIog) was constructing following the procedure described by Balbin-Tamayo et al.14 The reference and auxiliary electrodes were Ag/AgCl(KClsat) and a platinum wire, respectively.
Measures impedance spectroscopy were performed on a potentiostat/galvanostat EG & G Princeton Applied Research 263 coupled to a frequency response analyzer & G Princeton Applied EG Research 1025.
Experimental procedures
The immobilization of single-stranded DNA onto gold surfaces
Pre-treatment of the Gold Surface
Before each new series of measurements, the gold electrode was polished with 0,05 µm alumina suspension then the electrodes were sonicated in Milli-Q water during 15 min. Subsequently, to the electrode was applied to potential cycling from 0 V to 1,5 V vs. Ag/AgCl in 0,1 mol L-1 H2SO4 at a sweep rate (v) of 50 mV/s.13
The gold electrode was immersed in EtOH 65 % biotin 1 mmol L-1, Biotin-DNA 11,8 μmol L-1 for 16 h to prepare self-assembled biotin monolayer. The surface of the electrode before and after the coating was characterized cyclic voltammetry using a PalmSens instrument and Surface-Enhancement Raman Spectroscopy with micro-Raman model LabRAM-HR Horiba Jobin-Yvon (SERS) by Balbin-Tamayo et al.13
Electrochemical impedance spectroscopy (EIS) were used for electrochemical characterization of modified gold electrodes with self-assembled thiolated layers.
For bare and all modified electrodes, impedance spectra were recorded in the presence of 1mmol L-1 [Fe(CN)6]3-/4-, in 0,1 mol L-1 phosphate buffer solution at pH 6,9. EIS measurements were performed by applying a sine wave with amplitude of 10 mV (rms) on the open-circuit potential (EOC) value, and recording 10 points/frequency decade, in the 50 kHz-10 mHz frequency range. Before and after monolayer formation, the same electrode is subsequently used for impedance spectroscopy to assess impedance spectrum. Before recording the impedance diagram, all electrodes were submitted to the same stabilization program: 15 min at open circuit, followed by 15 min applying the relevant EOC value with measurement of the current until its stabilization.15 All experiments were repeated at least three times. From the impedance data, charge transfer resistance (Rct), was determinate using ZView2 software (Scribner Associates Inc.)
Hybridization with the complementary oligonucleotide was performed, after the biotin-DNA monolayer was formed for 16 hours, the modified electrode was contacted with complementary DNA solution of concentration 2,5 10-12 molL-1 in buffer. (0,1 molL-1 NaCl, Tris-Cl pH8, 0.1 molL-1 EDTA). For a minute at 95 °C (water bath), then 0 °C for one minute (ice bath) and at 65 ° C in a thermostated bath for one hour.11
The immobilization of single-stranded DNA onto epoxy-graphite electrode
Before each measurement, the electrode surface was moistened with water and then thoroughly smoothed, first using abrasive sandpaper and then, alumina paper in order to obtain a reproducible electrochemical surface.14 The surface of the electrode was characterized cyclic voltammetry and Raman Spectroscopy by Balbin-Tamayo et al.
Due to the high background resulting from nonspecific adsorption with noncovalent immobilization methods, covalent immobilization has been chosen to selectively link oligonucleotide probes to solid supports.16
The streptavidin was covalently linked to the surface carboxylic groups of the graphene oxide graphite-epoxy (EGoIIg) by peptide bonding 17, followed by binding of the biotinylated oligonucleotide to the streptavidin by the key-lock mechanism.18 Electrochemical detection was made of the oxidation signal of guanine of the oligonucleotide by square wave voltammetry.
Hybridization with the complementary oligonucleotide and proviral DNA was performed, after the biotin-DNA layer was formed, the modified electrode was contacted with complementary DNA solution of concentration 2,5 10-12 molL-1 in 0,1 mol L-1 phosphate buffer solution at pH 6,9 or clinical sample (proviral DNA HIV-1).
For a 5 minute at 95 °C, then 0 °C for minute and at 65 °C for one 25 minute in a PT-100 Programmable Thermal Controlled (MJ Research, Inc).11
Clinical samples
For preparation of the clinical samples, DNA from peripheral blood mononuclear cells was extracted from 100 μL of the whole blood by column affinity chromatography, using a QIAmp DNA mini Kit (Qiagen) and a QIAcube automated extraction kit. These samples were characterized in the Molecular Biology Laboratory of LISIDA, with direct detection of the viral load in peripheral blood using a standard system for in vitro diagnosis (COBAS Ampliprep/COBAS Taqman HIV-1 test v. 2.0) 19, which has a limit of detection of 20 copies mL-1.
Two samples with viral load <20 copies mL-1 were used as low positive controls, two samples with high viral load (≥104 copies mL-1) were high positive controls, and a negative control (0 copies mL-1) was also included.
Results and discussion
In the selective detection of the proviral DNA sequence of HIV-1, the length, composition and binding conditions such as pH, salt content, solvent and temperature were used under the experimental conditions accepted in the literature, such as pH, salt content, solvent and temperature to guarantee the specificity of the probe biotinylated oligonucleotide (Biotin-H1Gag1584).2,12 The specificity of the binding will increase with increasing temperature and decreasing salt content, which was taken into account during the hybridization process.12,20-22 The oligonucleotide probe used is 19 bases long, and the experimental condition followed the above criteria.
Hybridization onto gold surfaces
Electron transfer through the biotin layer was evaluated by electrochemical impedance spectroscopy (EIS) in the presence of complementary DNA and the redox marker ion Fe(CN)6 3-/-4 as an indicator of hybridization. Nyquist plots were fitted and the value of electron transfer resistance (Rct) was used to monitor the detection of hybridization of the target DNA, figure 1.
When the complementary oligonucleotide hybridized with the immobilized oligonucleotide on the surface of the electrode, the Rct value increased. An elongated sequence of HIV-1 proviral DNA was used as a sample, with a concentration of 2,5 10-12 mol L-1 and complementary to the self-assembled biotinylated single-stranded 19 base oligonucleotide on the surface of the gold electrode (table 1).
The increase in the Transfer Resistance (Rtc) value can be attributed to the electrochemical behavior of the completely complementary duplex. This must be attributed Hybridization with the target DNA, which introduces negatively charged phosphate groups on the electrode surface, increasing the electrostatic repulsion of the Fe (CN)6 3-/-4 ion detection layer. There is an increase in negative charge on electrode surface and charge transfer resistor, compared to probe DNA alone, table 1.23
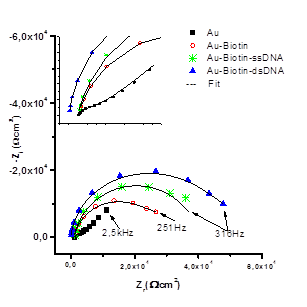
Fig. 1 Impedance spectra of potassium ferro/ferricyanide redox pare of: ■) bare Au electrode, ●) SAM Au-biotin, *) SAM Au-biotin-DNA and ▲) SAM Au-biotin-DNA/DNA complementary
Table 1- Values obtained for different SAMs on a gold electrode from impedance electrochemical spectroscopy, from the impedance adjustment, the data were determined using the ZView2 software.
| Samples | Rct / kΩcm2 | Coefficient of Variation % |
| Au | 2,0 ( 0,2 | 9,4 |
| Au-Biotin | 31,4 ( 0,5 | 1,6 |
| Au-Biotin-ssDNA | 42,0 ( 0,6 | 1,4 |
| Au-Biotin-dsDNA | 78,0 ( 0,6 | 7,5 |
Hybridization onto the graphene oxide graphite-epoxy electrode
In the graphene oxide modified epoxy-graphite electrode (EGIIog), streptavidin was covalently bound to the carboxyl groups on the surface of the EGIIog electrode by peptide bonding. The biotinylated oligonucleotide (Biotin-H1Gag1584) was then bound to streptavidin bound to the surface of the gr electrode by the keylock mechanism. Electrochemical detection of the electrode oxidation signal was performed at each step of the electrode surface modification, as shown in figure 1.
Figure 2 shows the square wave voltammograms obtained for the EGIIog electrode activated with NHS/EDC, EGIIog with covalently bound streptavidin, EGIIog with the biotinylated streptavidin/H1Gag1584 complex, and EGIIog with the biotinylated streptavidin/H1Gag1584 complex and the complementary oligonucleotide (c-DNA) in the supporting electrolyte (20 mmol L-1 phosphate buffer at pH 6,9). For the EGIIog electrode with the biotinylated streptavidin/H1Gag1584 complex, a potential signal was obtained at 0,94 V, which is close to the oxidation potential of guanine.24,25 Variation of this signal was used to detect DNA hybridization.
The carboxyl groups of the graphene oxide-modified graphite-epoxy electrodes enabled immobilization of the biotin-H1Gag1584 oligonucleotide on the electrode by the key-lock mechanism involving the biotinylated oligonucleotide and streptavidin. This immobilization avoids non-specific adsorption, allows a good spatial configuration of the oligonucleotide and guarantees a selectivity of the modified electrode.26 This form of immobilization is an important element in the reliable and sensitive detection of HIV-1 proviral DNA (the analyte) from hybridization.
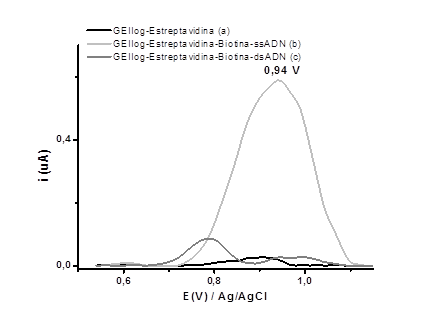
Fig. 2 Square wave voltammograms obtained in phosphate buffer at pH 6.9 using the GEIIog electrode with (a) streptavidin, (b) streptavidin-biotin-H1Gag1584, and (c) streptavidin-biotin-H1Gag1584/c-DNA
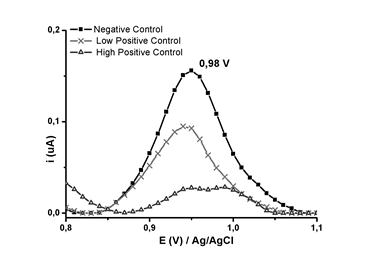
Fig. 3 Square wave voltammograms for the negative (-■-), low positive (-x-), and high positive (-(-) controls, obtained in 20 mmol L-1 PB (pH 6,9)
The signal of the voltammogram at 0.98 V (Figure 3) could be attributed to the oxidation of guanine in the biotinylated oligonucleotide H1Gag1584. The intensity of the signal current at 0,98 V decreased when the biosensor (the electrode with the oligonucleotide) came into contact with the positive controls (complementary) under experimental hybridization conditions.27 The decrease in current at this potential is attributable to the formation of hydrogen bonds between the complementary bases of the biotinylated oligonucleotide and the proviral DNA, which resulted in a decrease in electron density.28,29 The reason why hybridization can be detected for a DNA sample at a potential of 0.98V.
To evaluate the reproducibility of the electrodes (n = 10), negative, low positive and high positive controls were used, with values of 0,197 ± 0,04, 0,119 ± 0,02 and 0,036 ± 0,01 obtained, respectively. The results revealed appreciable differences between the negative, low positive (less than 20 RNA molecules mL-1), and high positive (105 RNA copies mL-1) controls at 0,98 V. Although quantitative correlations between the viral load and the amount of proviral DNA has not been established, it is known that more than one virion can infect the same cell, and that more than one region of proviral DNA may be inserted into the chromosomal DNA. The amount of proviral DNA detectable in samples with high viral loads may be influenced by a greater number of infected cells having more than one proviral DNA region, which would be in agreement with the marked decrease of the signal for the high positive control. In the case of samples with low viral load (such as the low positive control), the signal is only due to proviral DNA regions in the few infected cells. Decreasing the intensity of the guanine oxidation current for the biotin-H1Gag1584 oligonucleotide at a potential of 0,98 V enabled the detection of proviral HIV-1 DNA by square wave voltammetry in phosphate buffer at pH 6,9.
Detection of proviral DNA can reduce the post-exposure window period to about 11 days, and so may also be used to detect HIV-1 in individuals who have not yet presented an immune response, or to resolve indeterminate serology results.
Conclusions
On the biotin-H1Gag1584 oligonucleotide modified gold electrode, the difference in Rtc values indicated that it exhibited a sensitive response to the detection of hybridization of a complementary DNA probe without markers. Consequently, successful and tag-free detection of HIV-1 ssDNA is demonstrated. While the biotin-H1Gag1584 oligonucleotide modified epoxy graphite oxide composite electrode offers the possibility of covalent immobilization of specific oligonucleotides. Square wave voltammetry allowed the successful detection of HIV-1 proviral DNA derived from chromosomal DNA of whole blood lymphocytes in clinical samples; by decreasing the intensity of the guanine oxidation current of the biotin-H1Gag1584 oligonucleotide to a potential of 0,98 V. The results obtained in this work showed the suitability of the biotin-H1Gag1584 oligonucleotide for two electrochemical detection systems of HIV-1 tag-free proviral DNA.















