My SciELO
Services on Demand
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista de Protección Vegetal
Print version ISSN 1010-2752
Rev. Protección Veg. vol.27 no.3 La Habana Sept.-Dec. 2012
ORIGINAL ARTICLE
Identification of new hosts for Ralstonia solanacearum (Smith) race 2 from Colombia
Identificación de nuevos hospedantes de Ralstonia solanacearum (Smith) raza 2 en Colombia
Jorge Prieto RomoI, Juan Gonzalo Morales OsorioII, Mauricio Salazar YepesIII,*
IAgronomical Engineer, Universidad Nacional de Colombia sede Palmira, Palmira-Colombia. E-mail: jorgeprieto150@yahoo.com.
IIProfessor, IA, MSc, PhD, Universidad Nacional de Colombia, sede Medellín, Facultad de Ciencias Agropecuarias, Departamento de Ciencias Agronómicas, Colombia. E-mail: jgmoraleso@unal.edu.co.
IIIProfessor, IA, MSc, PhD, Universidad Nacional de Colombia, sede Medellín, Facultad de Ciencias, Escuela de Biociencias, Colombia.
ABSTRACT
Ralstonia solanacearum Smith induces the Moko disease on banana, plantain and heliconia flowers. The objectives of this work were: I) to identify R. solanacearum hosts in weeds or cultivated hosts and II) to determine its pathogenicity on the susceptible host plantain cv. Dominico-Hartón. A survey in search of natural hosts of R. solanacearum race 2 was performed in selected Colombian regions. Sixty bacterial colonies showing R. solanacearum characteristics in a semi-selective medium were used in further pathogenicity tests in the susceptible plantain plants. Twenty six isolates induced Moko disease symptoms in plantain during the 60 days of evaluation. Twelve new hosts were found for R. solanacearum at the worldwide level: nine of them were weeds (Euphorbia graminea Jacq., Blechum piramidatum Lam., Oxalis latifolia Kunth, Cuphea micrantha Kunth, Eleusine indica L., Gliricidia sepium Kunth ex Steud., Lobelalia xalapensis Kunth, Stachys lamioides Benth., Salvia aff. lasiocephala Hook. & Arn.) and three cultivated crops (Colocasia esculenta L., Cucurbita maxima Duchesne and Psidium guajava L.). The presence of R. solanacearum race 2 in weeds and cultivated crops should be managed as an important component of an integrated Moko disease control program.
Key words: Musa spp., Ralstonia solanacearum, Moko disease.
RESUMEN
Ralstonia solanacearum Smith ocasiona la enfermedad denominada Moko en cultivos de banano, plátano y heliconias. Los objetivos de este trabajo fueron: I) identificar hospedantes de R. solanacearum en malezas o plantas cultivadas y II) determinar su patogenicidad en la variedad de plátano susceptible Dominico-Hartón. Se realizó un muestreo para identificar hospedantes naturales de R. solanacearum raza 2 en regiones de Colombia. Sesenta colonias aisladas en medio semi-selectivo mostraron características típicas de R. solanacearum y fueron usadas posteriormente para la evaluación de patogenicidad en plantas susceptibles de plátano. Veintiséis cepas indujeron los síntomas típicos de la enfermedad del Moko en las plantas de plátano, durante el período de evaluación de 60 días. Se encontraron doce hospedantes no informados previamente a nivel mundial para R. solanacearum: nueve fueron malezas (Euphorbia graminea Jacq., Blechum piramidatum Lam., Oxalis latifolia Kunth, Cuphea micrantha Kunth, Eleusine indica L., Gliricidia sepium Kunth ex Steud., Lobelalia xalapensis Kunth, Stachys lamioides Benth., Salvia aff. lasiocephala Hook. & Arn.) y tres correspondieron a hospedantes cultivados )Colocasia esculenta L., Cucurbita maxima Duchesne y Psidium guajava L.). La presencia de R. solanacearum raza 2 en malezas y hospedantes cultivados debe ser manejado como un componente importante en el programa integrado de control de la enfermedad del Moko.
Palabras clave: Musa spp., Ralstonia solanacearum, enfermedad del Moko.
INTRODUCTION
The plant pathogenic bacterium Ralstonia solanacearum Smith has a wide host range infecting more than 200 different plant species within 53 taxonomic families (1, 2). There are five different races and an equal number of biovars of R. solanacearum depending on the host range and the biochemical and physiological characteristics, respectively. Race 1 mainly infects cultivated plants belonging to the Solanaceae family and diploid bananas; whereas race 2 causes the Moko disease (3), arguably the most important bacterial disease in triploid bananas, plantain and the ornamental plants heliconias in the tropics. Race 3 attacks potato, tomato and geranium; race 4 has been reported causing disease on ginger (4, 5, 6, 7) and race 5 (biovar 5) is specialized on Morus (7).Most susceptible hosts include cultivated plants within the Solanaceae family (tomato, potato, tobacco, bell pepper), Fabaceae (peanut), Musaceae (banana and plantain), and many others (2, 8). Previous studies aiming to determine R. solanacearum natural hosts have demonstrated the bacteria ability to asymptomatically colonize survive and persist in the absence of susceptible plants (1, 4, 9).
In Colombia, 17 new host weeds for races 1, 2 and 3 were identified (4); on the other hand, Granada (10) determined that roots from common weeds found in plantain crops might be asymptomatic hosts for R. solanacearum race 2. Meanwhile in Honduras and Costa Rica, several hosts other than banana have been identified in natural ecosystems (5). More than 50 % of R. solanacearum host species reported have been weeds (4). R. solanacearum can be spread very fast by water streams, machinery, insects, seeds, labour tools, crop workers and animals (1, 4, 6, 11). Colombia is the second larger plantain producer and the third largest banana exporter in the world, besides many wild heliconia species are native from Colombia, and Moko disease is a permanent threat to these current and potential crops.
The disease was first reported in Tolima department, Colombia, in1954 by Galvez and Lozano (12), and from this place, it spread throughout the country. As a consequence of Moko spreading, losses could reach 100% of production if the treatments were not correctly and promptly applied. In the most important Colombian banana growing area (Urabá), more than 611 hectares have been destroyed at an average rate of 16-17 hectares per annum (13). Additionally, in banana growing areas, the control and quarantine tactics for Moko have a high economic impact since nine plants must be destroyed per each infected plant (11) and a six month quarantine established (13) in the area.
Despite the numerous investigations on R. solanacearum, many aspects of its ecology, survival, spread and host range in the tropics are still poorly understood.
The objectives of this work were I) to identify R. solanacearum hosts on weeds or cultivated plants and II) to determine its pathogenicity on the susceptible host plantain cv. Dominico-Hartón.
MATERIALS AND METHODS
Survey and sample collection
Bacterial isolates were obtained from weeds and cultivated plants in the crops of banana, plantain, heliconia, bell pepper, potato and tomato growing in areas of the three Colombian Departments named Caldas, Quindío and Valle del Cauca (Figure 1). Special attention was given to those fields with a previous report of Moko disease. Plants were sampled within or around Moko foci including symptomless plants. Plants showing Moko symptoms were collected;fields with apparently healthy crops were also randomly sampled. Plant genera and species were identified with the assistance of the Herbarium staff from Universidad Nacional de Colombia (Medellín and Palmira Headquarters), who provided a list of plant scientific names.
Identification of R. solanacearum from different host plants
Bacteria were isolated and identified in the molecular biology laboratory at Universidad Nacional de Colombia headquarters Palmira, using the below described protocols.
Individual plants were surface sterilized and root and stem vascular tissues were extracted with a new sterile scalpel blade. Small pieces of the extracted tissues (1-2mm2) were placed on Kelman-triphenyltetrazolium chloride (TZC) TZC semi-selective medium and incubated (14). Individual virulent mucoid colonies with pink centers appeared after 48-60 hours, then developed blood red whorls showing a reddish-golden halo. Bacterial colonies showing R. solanacearum distinctive features were further identified using the ImmunoStrip® test Kit from Agdia® for specific detection of R. solanacearum, following the manufacturer´s instructions.
Colonies confirmed as R. solanacearum were grown overnight in liquid semi-selective medium from South Africa (SMSA) at 28°C under constant shaking (250rpm). After incubation, glycerol was added up to 30% and the strains kept at -80°C for further use.
Pathogenicity test
The isolates obtained were inoculated on Moko susceptible seedling plants cv. Dominico-Hartón. For this study, plantain seedlings were obtained from a disease-free certified producer farm and grown in the greenhouse for pathogen containment in 3kg pots with a sterile soil-organic matter substrate (3:1). After one month, 15g of a mixture of nitrogen and potassium (3:2) were applied. The plantlets were grown for another month under similar conditions before inoculation.
Each strain was inoculated in each group of four individual two month old plantain plants. A completely randomized block design with four replicates was used as experimental model. Each plant was injected with a syringe in the pseudostem at a height of 15cm from soil with 2,5 ml of a solution containing each individual strain at a concentration of 108 colony forming units (CFU).mL-1. Plants inoculated with liquid medium and plants non-inoculated were used as negative controls. After inoculation plants and controls were grown at 27°C and 70% relative humidity in a greenhouse with an irrigation system for watering plants every two days.
Moko disease symptoms were recorded every three days after inoculation (DAI) during 60 days. A strain was considered positive when clear Moko symptoms were induced on at least three plants per inoculated strain (12).
Bacteria were re-isolated from diseased plants in Kelman semi-selective medium to confirm Koch´s postulates. Disease severity was scored by visual evaluation according to the semi-quantitative scale reported by Obregón et al. (12) (Table 1).
Every individual strain was considered as a different treatment. Disease severity was estimated by the area under the disease progression curve (AUDPC) for each bacterial strain, represented as the sum of the area of the polygons for each evaluation as described by Campbell and Madden (15). An analysis of variance of the AUDPC values was performed; means were square root transformed and compared by the Least Significant Difference (LSD) multiple range test (P<0.05). Differences were calculated from 4 repetitions for each treatment.
RESULTS
Identification of R. solanacearum from different host plants
Using Kelman´s semi-selective medium, sixty strains were selected by the characteristics previously reported by Kelman (14) for Pseudomonas solanacearum (currently R. solanacearum) such as large, elevated, fluidal, and either entirely white or with a pale red center colonies (Table 2a, Table 2b cont., Table 2c cont.). All strains were confirmed as R. solanacearum by serological studies.
Forty one strains were obtained from the central Colombian department Quindío, sixteen from Valle del Cauca, and three from Caldas. Eighteen strains were from cultivated plants such as plantain (three), tomato (five), potato (three), bell pepper (two), heliconia flower (one), guava (one), mandarin (one) and squash (one) and other forty two strains from non-cultivated plants (Table 2a, Table 2b cont., Table 2c cont.).
Pathogenicity test
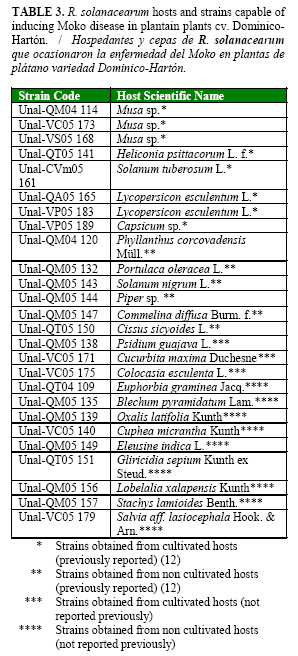
From the sixty strains inoculated on plantain plants, only twenty six (from cultivated and non-cultivated hosts) were capable of producing Moko disease (Table 3). Interestingly, other 27 strains developed initial Moko symptoms such as mild wilt (degree 1) and yellowing (degree 2) (Table 1), but most of the plants recovered during the evaluation period, and only two or less plants per strain inoculated remained affected (Data not shown).
First wilting symptoms were observed between 12 to 18 DAI. At eighteen DAI, the plants showed yellowing. At thirty DAI, the typical Moko symptoms such as necrosis of the youngest leaf and yellowing of 2-3 youngest leaves were recorded (Figure 2). The first plant dead appeared forty two DAI. At forty five DAI, twenty one isolates induced Moko wilted plant symptoms with a scale value between 1 and 5 (Figure 2).
Strains (Unal-QA05 165) and (Unal-VP05 183) from tomato and strain (Unal-VP05 189) from bell pepper induced Moko symptoms and caused plant death in at least one plant out from four inoculated per strain. Strain (Unal-CVm05 161) from potato induced symptoms showing scale values between 2 and 4 but did not cause death to inoculated plants. Two strains were obtained from Cucurbita maxima and one of them was pathogenic to plantain (Unal-VC05 171). All Moko-positive strains showed R. solanacearum characteristics described by Kelman (14) when re-isolated in semi-selective medium confirming Koch´s postulates for these 26 strains.
Positive strains for Moko disease were obtained from twelve new hosts not reported previously at the worldwide level: E. graminea, B. piramidatum, P. guajaba, O. latifolia, C. micrantha, E. indica, G. sepium, L. xalapensis, S. lamioides, C. esculenta, C. maxima, S. aff. lasiocephala, from which C. maxima, C. esculenta and P. guajava are cultivated hosts.
The AUDPC calculated for 26 positive strains showed different levels of aggressiveness on plantain cv. Dominico-Hartón (Figure 3). The strains Unal-QM05 138 (from P. guajava) and Unal-QM05 147 (from C. diffusa), developed Moko disease slowly compared with most strains that produced AUDPC values similar to strain Unal-VS05 168 original from plantain (Figure 3). Strains 135 and 149 from weeds and 161, 165, 168, 189 from cultivated hosts, grouped together with the highest AUDPC values (Figure 3, Table 2a, Table 2b cont., Table 2c cont.).
DISCUSSION
An effective management of the Moko disease requires knowledge about R. solanacearum host range, because several plants may act as inoculum reservoirs for important cultivated crops like banana and plantain.
In this work, twelve new hosts for R. solanacearum race 2 were described in areas where banana, plantain, heliconias, bell pepper, potato and tomato are grown in Colombia . More than 200 plant species distributed in about 53 families have been reported as hosts for R. solanacearum (1, 16). In a recent work, we reported eight new R. solanacearum race 2 hosts: C. nutans, S. cinerea, T. glandulosa, P. hirtus, P. pellucida, T. cumanensis, Desmodium sp. and C. sicyoides. The latter (C. sicyoides) is classified in the botanical family Vitaceae, which any of its members had previously been reported as host for R. solanacearum race 2 (12).
A recent molecular characterization of Colombian strains virulent on Musa sp. showed that this population was sub-structured (Fst=0,66) with the host as the main factor of differentiation (17).
Although several works have been performed, the basic biology and ecological interactions between R. solanacearum, its different hosts and the environment are not completely clear. Our results together with previous reports suggest that R. solanacearum has an extraordinary wide host range, aggressiveness, environment adaptation ability and ability to survive in an asymptomatic, symptomatic, systemic or endophytic way (1, 16). The host range for R. solanacearum race 2 is wider than the expected one and the bacteria, pathogenic to plantain cv. Dominico-Hartón, can be found in several regions and diverse ecosystems even different from those in which the plantain or related crops like bananas are frequently found.
The results obtained through this work and other reports support the hypothesis that R. solanacearum exhibits an endophytic phase during its life cycle. This phase is important for bacteria survival and renovation of soil and host populations (1). Interestingly, a strain from potato collected at high altitude (3000 meters above mean sea level (m.s.l)) was pathogenic to plantain cv. Dominico-Hartón, which is usually cultivated below 2000 m.s.l. This finding reflects the remarkable adaptability of this plant pathogen to different environments, soil types, moisture conditions, hosts and temperatures.
The presence of host weeds in crop fields should be considered for disease prevention and management. Host range is determined by a number of different factors which are not completely understood. Some strains such as GMI1000, originally isolated from tomato in French Guyana, have been reported to have a broad host range, including different botanical families, whereas other strains have exhibited a narrower host range (8). Genin (8) pointed out that research on R. solanacearum host range has two important limitations to be considered. First, some strains are referenced only from the host they were originally isolated and second, artificial inoculations may overestimate the natural host range.
In our work we found twelve hosts that might have implications in crop disease management. Agricultural practices introduce tissue wounds which help bacterial colonization in a similar way artificial inoculations do and should be considered in disease control. Asymptomatic infections are a natural reservoir of inocula that contribute to spread the Moko disease.
R solanacearum disease induction is a complex process which involves environmental and crop conditions, the different hosts and a number of pathogenicity determinants from the bacterium, named effector proteins (18, 19). Penetration and colonization of host tissues by the pathogen is a required step for disease induction. Plants exhibit non-host resistance against most potentially pathogenic microbes which delimits host range. The nature of this resistance may involve preformed physical or chemical barriers or induced defense responses activated by microbe associated molecular patterns (MAMP´s), damage associated molecular patterns (DAMP´s) or pathogen effector proteins. In addition, R. solanacearum may colonize some hosts asymptomatically making host range determination a complex scientific challenge. Unlike most phytopathogenic bacteria, which need natural or wound induced openings, some R. solanacearum strains are able to invade plants through intact roots (8). Wounds produced during agricultural labors, insect damage or artificial inoculations may help overcome natural barriers against plant tissue infection. Molecular determinants of host range are beginning to be elucidated (18, 19). Some recent studies reported that some R. solanacearum type 3 secreted effectors may determine host range as exemplified by the AvrA and the popP1 genes from the GMI1000 strain which confer specificity on tobacco (20). It is expected that during the upcoming years knowledge will increase to comprehensively understand factors and mechanisms determining R. solanacearum host range (19, 21).
R. solanacearum race 3 induces disease mainly on potato and tomato and other solanaceous crops and weeds in the higher elevations of the tropics. In this work, we obtained two strains from tomato (Unal-QA05 165, Unal-VP05 183) and one from bell pepper (Unal-VP05 189) which induced mild Moko disease and caused plant death in artificially inoculated plantains. In a previous work, we observed similar results with one strain from tomato which induced mild disease symptoms in plantain (12). A similar finding was reported by Buddenhagen (5) in seed-bearing Musa sp. Belalcázar et al. (4) obtained mild symptoms on Physalis peruviana inoculated with strains from race 1, 2 and 3 and reported P. peruviana as R. solanacearum resistant. Buddenhagen (5) reported wilt symptoms in tomato and Physalis angulata L. inoculated with R. solanacearum strains isolated from abandoned plantain and heliconia plantations in Costa Rica.
According to French (6), the host range under field conditions is well defined; bacteria may colonize tissues but do not induce disease, which may not reflect completely the situation under experimental conditions. For example, it is possible that structural penetration barriers, which may determine host range and the first defense mechanism, may be surpassed by bacterial artificial injection. Further evidence was provided by Prior and Fegan (22), who reported ecotypes of R. solanacearum race 2 inducing characteristic Moko symptoms on Musa spp. under field conditions and having the ability to be pathogenic on tomato and other solanaceous plants when the bacteria were stem inoculated; however, these strains have not been isolated from in-field wilted solanaceous plants. According to French (6), race 3 is found mainly at high latitudes north or south or at high altitude in the tropics. In Colombia, race 3 is endemic at altitudes higher than 2200 m.s.l. French and Gutarra (23) reported that R. solanacearum race 3 was able to adapt in-vitro to a warm environment. These reports may indicate a possible explanation for our results. Furthermore, strains classified within race 1 are widely distributed in the tropics and may infect solanaceaous plants and diploid bananas (7), showing the ability of one R. solanacearum race to infect plants from different families. The results achieved in this work may be explained by a strain with pathogenicity to both tomato and plantain, or by a favorable environment for the pathogen under the conditions used in this experiment. It is important to consider that the experimental conditions used in this work may reflect what could happen under agricultural conditions where labors and inappropriate tool used may cause plant tissue injuries facilitating pathogen entry, or by roots pest damage.
The fact that AUDPC values observed for most strains were similar to strain Unal-VS05 168 obtained from plantain, could indicate the potential pathogenicity of R. solanacearum strains from hosts different to those obtained from Musa sp. This possibility should be considered for crop disease management to prevent bacteria spread.
Belalcazar et al. (4) proposed to eradicate weed and cultivated hosts from quarantined areas to make an effective Moko control. This measure has proven effective to diminish Moko incidence in large areas like Urabá in Colombia (13). Moko control includes a six month quarantine period on a 5m radius around disease foci and permanent weed control. Crop rotation may be an alternative for this long quarantine period, but the increasing R. solanacearum race 2 host range must be an important factor to consider because symptomless plants colonized by R. solanacearum may be a source of inoculum for banana or plantain re-infection after the above mentioned quarantine period (12).
Squash (C. maxima) is frequently used in the Colombian regions Quindío and Valle del Cauca for crop rotation after banana or plantain eradication during Moko control. In this work, two R. solanacearum strains were isolated from C. maxima plants and one was positive for Moko induction in plantain cv. Dominico-Hartón suggesting that squash was not appropriate as a rotation crop in a banana or plantain Moko eradication program.
Bacteria diversity has been classified into four molecular phylotypes which maybe arose by geographical isolation (22). These phylotypes show little or no correlation between host range and phylogenetic relationships (8). Wicker et al. (24) reported a group of strains, some of them obtained from plants of the family Cucurbitaceae, belonging to phylotype II / sequevar 4 (II/4). Although they clustered with the group of Moko disease strains, they were not pathogenic to banana. Interestingly, strains from family Cucurbitaceae were able to asymptomatically infect plantain (cooking banana) cv. Dominico-Hartón (24).
It is very important to continue surveys to determine new natural hosts and the role they play in bacteria dispersion. Host range, particularly asymptomatic hosts, is one factor that, with many others such as soil and debris bacteria persistence and survival, early diagnosis, effective quarantine period, crop workers education, and inoculum dispersion, should be studied in more detail for a better Moko disease control and eradication.
CONCLUSION AND PERSPECTIVES
Findings in this work suggest an insufficient knowledge of R. solanacearum host range. Surveys in search of an elucidation of a comprehensive host range should be continued. As cultivated and not cultivated hosts other than banana, plantain and heliconias may be inoculum sources for disease spread, host management should be considered within a Moko disease control and eradication program.
ACKNOWLEDGEMENTS
Authors would like to express acknowledgements to SENA-COLCIENCIAS for project funding (project: Development of a method of asymptomatic detection of the banana and plantain Moko disease PCR-based and applications to disease management, code: 8242-07-16025), to Universidad Nacional de Colombia sede Palmira Research Office (DIPAL) and to Herbarium MEDEL for the taxonomic plant identification. The funding source(s) has no involvement in the conduct of the research, preparation of the paper, the study design, the collection, the analysis and interpretation of data, the writing of the report or in the decision to submit the paper for publication.
REFERENCES
1. Hayward AC. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol. 1991;29:65-87.
2. Rodrigues L, Destéfano S, Diniz M, Comparoni R, Rodrigues-Neto J. Pathogenicity of Brazilian strains of Ralstonia solanacearum in Strelitzia reginae seedlings. Tropical Plant Pathology. 2011;36(6):409-413.
3. Morales JG, Castañeda D. Ralstonia solanacearum, otro nombre para Pseudomonas solanacearum, el agente causal del Moko. Carta Inf Augura. 2001;211(1):4-6.
4. Belalcazar S, Rosales F, Pocasangre L. El «Moko» del plátano y banano y el rol de las plantas hospederas en su epidemiología. In XVI Reunión Internacional de ACORBAT, 26 of September-1 Octubre, Oaxaca, México. Publicación especial; 2004. p. 16-35.
5. Buddenhagen IW. Bacterial wilt of certain seed-bearing Musa spp caused by tomato strain of Pseudomonas solanacearum. Phytopathology. 1962;52(3):286.
6. French ER. Interaction between strains of Ralstonia solanacearum, its hosts and the environment. In Persley, G.J. (Ed.), Bacterial wilt disease en Asia and the South Pacific. ACIAR Proceedings, vol. 13. Los Baños, Philiphines; 1985.p. 99-104.
7. European Plant Protection Organization (EPPO). Ralstonia solanacearum. OEPP/EPPO Bulletin. 2004;34:173-178.
8. Genin S. Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytologist. 2010;187:920-928.
9. Cruz L, Eloy M, Quirino F, Oliveira H, Tenreiro R. Molecular epidemiology of Ralstonia solanacearum strains from plants and environmental sources in Portugal. European Jour Plant Pathology. 2012;133(3):687-706.
10.Granada G. Manejo integrado del Moko del plátano (Pseudomonas solanacearum) bajo condiciones de la zona cafetera del Quindío. In Tecnología del eje cafetero para la siembra y explotación rentable del cultivo del plátano. Comité departamental de cafeteros del Quindío. Tercer informe técnico, 1994-1996, Regional 9, CORPOICA, Armenia, Colombia; 1996. p. 95-96.
11.Munar-Vivas O, Morales-Osorio J, Castañeda-Sánchez D. Use of field-integrated information in GIS-based maps to evaluate Moko disease (Ralstonia solanacearum) in banana growing farms in Colombia. Crop Protection. 2010;29:936-941.
12.Obregón M, Rodríguez P, Morales J, Salazar M. Hospedantes de Ralstonia solanacearum en plantaciones de banano y plátano en Colombia. Rev Fac Nal Agr Medellín. 2008;61(2):4518-4526.
13.Castañeda D, Espinosa J. Comportamiento e impacto de la enfermedad de Moko en la zona de Urabá (Colombia), en las últimas tres décadas y media y propuesta de un índice de riesgo de la enfermedad. Rev Fac Nal Agron Medellín. 2005;58(1):2587-2599.
14.Kelman A. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathology. 1954;44(12):693-695.
15.Campbell CL, Madden LV. Introduction to Plant Disease Epidemiology. John Wiley & Sons, New York City, 1990. 532p.
16.Kumar A, Prameela T, Suseela Bhai R, Siljo A, Biju N, Anandaraj M, et al. Small cardamom (Elettaria cardamomum Maton.) and ginger (Zingiber officinale Roxb) bacterial wilt is caused by same strain of Ralstonia solanacearum: a result revealed by multilocus sequence typing (MLST). Eur J Plant Pathol. 2012;132:477-482.
17.Cardozo C, Rodríguez P, Cotes J, Marín M. Variabilidad genética de la bacteria Ralstonia solanacearum (Burkholderiales: Burholderiaceae) en la zona bananera de Urabá (Colombia). Rev Biol Trop. 2010;58(1):31-44.
18.Genin S, Denny T. Pathogenomics of the Ralstonia solanacearum Species Complex. Annu Rev Phytopathol. 2012;50:4.1-4.23.
19.Remigi P, Anisimova M, Guidot A, Genin S, Peeters N. Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytologist. 2011;192:976-987.
20.Poueymiro M, Cunnac S, Barberis P, Deslandes L, Peeters N, Cazale-Noel A, et al. Two Type III Secretion System Effectors from Ralstonia solanacearum GMI1000 Determine Host-Range Specificity on Tobacco. Molecular Plant-Microbe Interactions. 2009;22(5):538-550.
21.Wicker E, Lefeuvre P, de Cambiaire JC, Lemaire C, Poussier S, Prior P. Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. ISME J. 2012;6(5):961-74.
22.Prior P, Fegan M. Recent developments in the phylogeny and classification of Ralstonia solanacearum. Acta Horticulturae (ISHS). 2005;695:127-136.
23.French E, Gutarra L. Adaptabilidad de Pseudomonas solanacearum razas 1 y 3 al frío. In: Resúmenes del II Congreso Nacional de Investigadores Agrarios del Perú, 12-16 de Agosto, 1974, Lima, Perú; 1974. p. 44.
24.Wicker E, Grassart L, Coranson-Beaudu R, Mian D, Guilbaud C, Fegan M, Prior P. Ralstonia solanacearum strains from Martinique (French West Indies) exhibiting a new pathogenic potential. Applied and Environmental Microbiol. 2007;73(21):6790-6801.
Recibido: 27-1-2012.
Aceptado: 24-4-2012.
*Author for correspondence: Mauricio Salazar Yepes. Professor, IA. MSc, PhD, Universidad Nacional de Colombia, sede Medellín, Facultad de Ciencias, Escuela de Biociencias. E-mail: masalazay@unal.edu.co