My SciELO
Services on Demand
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Biotecnología Aplicada
On-line version ISSN 1027-2852
Biotecnol Apl vol.29 no.1 La Habana Jan.-Mar. 2012
RESEARCH
Predicted proteins of Neisseria meningitidis as potential vaccine candidates: from in silico analyses to experimental corroboration
Proteínas predichas de Neisseria meningitidis como posibles candidatos vacunales: de los análisis in silico a la corroboración experimental
Darien García1, Daniel Yero1,2, Olivia Niebla1, Karem Cobas1, Yasser Perera1, Evelin Caballero1, Maite Delgado1, Rolando Pajón1,3
1Meningococcal Research Department, Division of Vaccines, Center for Genetic Engineering and Biotechnology, CIGB. Ave. 31, Cubanacan, PO Box 10 600, Havana, Cuba.
2Department of Molecular Biology, Division of Biotechnology, Finlay Institute. Ave. 27, La Lisa, PO Box 11 600, Havana, Cuba.
3Faculty of Medicine, University of Calgary, Heritage Medical Research Building, Hospital Drive NW, Calgary, Canada.
ABSTRACT
Neisseria meningitidis serogroup B infections are a serious health threat to the world that cannot be prevented by vaccination. Here, we report an analysis of the MC58 Neisseria meningitidis genome aimed at the identification of new potential vaccine candidates. ‘Hypothetical’ and ‘conserved hypothetical’ annotated genes, together with those with putative functions related to the cell envelope, were subjected to extensive sequence similarity searches, as well as motif, cellular location, and domain analyses complemented with manual curation. As a result, a set of 35 uncharacterized ORFs, predicted to encode for surface exposed or virulence related proteins, was identified. The candidates were subdivided in three categories: 1) predicted outer membrane proteins (OMPs) unique of the Neisseria genus; 2) conserved OMPs from various genus and 3) proteins homologous to known OMPs or to proteins previously found to be immunogenic in animal models. Two of the final candidates, nmb1126 and nmb0181, were cloned and expressed in Escherichia coli. The resulting products were purified by Metal Chelating Chromatography and used to immunize mice. The recombinant proteins were capable of inducing antibodies against the native antigen in preparations of a panel of three strains and displayed bactericidal activity against the homologous strains.
Keywords: Neisseria, in silico analyses, open reading frames, antigen, vaccine.
RESUMEN
Las infecciones producidas por el serogrupo B de Neisseria meningitidis constituyen un serio problema de salud mundial que no puede ser prevenido mediante la vacunación. En este artículo reportamos un análisis del genoma de la cepa MC58 de N. meningitidis desarrollado con el objetivo de identificar nuevos posibles candidatos vacunales. Los genes anotados como ‘Hipotéticos’ e ‘Hipotéticos conservados’, en conjunto con aquellos que pudieran presentar funciones relacionadas con la envoltura celular, estuvieron sujetos a extensos análisis de similitud de secuencia, localización celular y de motivos y dominios complementados con curación manual. Como resultado, 35 marcos de lectura abiertos no caracterizados, predichos para codificar proteínas de superficie o relacionadas con la virulencia fueron identificados. Los candidatos fueron subdivididos en tres categorías: 1) proteínas de membrana externa (PME) únicas del género Neisseria; 2) PMEs conservadas en varios géneros y 3) proteínas homólogas a PME conocidas o a proteínas previamente reportadas como inmunogénicas en modelos animales. Dos de estos candidatos, el nmb1126 y el nmb0181 fueron clonados y expresados en Escherichia coli. Los productos resultantes fueron purificados por cromatografía de afinidad a quelatos metálicos y empleados para inmunizar ratones. Las proteínas recombinantes fueron capaces de inducir anticuerpos que reconocieron el antígeno nativo del meningococo en un panel de tres cepas y que mostraron actividad bactericida contra la cepa homóloga.
Palabras clave: Neisseria, análisis in silico, sistemas de lectura abierta, antígeno, vacuna.
INTRODUCTION
Neisseria meningitidis is the primary cause of bacterial meningitis worldwide. Despite the available antibiotics, there is 10% mortality on a global basis and convalescent patients may have serious sequels [1]. Serogroups A, B, C, W135 and Y are responsible for around 95% of the cases worldwide, although serogroup B shows the highest incidence in developed countries [2].
Although a polysaccharide vaccine has been successfully developed against several serogroups this strategy has not been feasible for serogroup B, because of the marked structural homology between its capsular polysaccharide and human neural antigens [3].
Some alternatives, such as the application of vaccines based on meningococcal outer membrane vesicles (OMV) have proved successful in controlling disease outbreaks [4, 5]. However, despite this partial success, these vaccines do not confer optimal protection against the broad panel of circulating strains, due to the variability of the major antigenic proteins [4-6]. Therefore, the search for conserved subcapsular proteins capable of inducing a broad and protective immune response has become an important objective.
Alternative to traditional laboratory tools, the complete genome sequencing of different strains of N. meningitidis have been an accelerator to vaccine candidates study allowing the employment of technologies like proteomics and immunoproteomics (for a review see [7]. Also reverse vaccinology emerges as a powerful new approach for the identifi cation of novel potential vaccine antigens. Its application can significantly reduce the time and cost required to achieve these goals. [8]. After the availability in 2002 of the genome sequence of the strain MC58 serogroup B N. meningitidis, Pizza et al. introduced a novel methodology to predict outer membrane proteins (OMPs) with potentiality to confer protection against meningococcal infection [9]. Several antigen candidates were identified in this study. Five of them were included in a multicomponent vaccine currently in a phase III clinical trial [10]. Some of the antigens included in this preparation showed extensive sequence variability and, therefore, they induced an immune response against a limited spectrum of strains. However, by combining these proteins in a multicomponent vaccine, they have been able to generate a broadly crossreactive immune response [9, 11-13].
Although the work of Pizza et al. [9] was pioneer in this field, many other investigators have applied variations of this method, combined with other experimental tools to the screening for new antigen candidates in Neisseria and other microorganisms. In this regard, different software combinations, including in-house scripts, have been used in conjunction with microarrays, sucrose gradients, and fusion screens among others, in the search for outer membrane proteins or molecules with potential immunogenic value [14-18].
In this article we describe an alternative bioinformatics strategy using free internet servers to analyze the genome of the same N. meningitidis strain that was processed by Pizza et al. We focused on hypothetical open reading frames (ORFs) and genes with putative functions related to the bacterial envelope. The degree of conservation of the putative antigen was the fundamental criterion for selection. The analyses of orthologous groups, motifs, domains and genetic neighborhood were equally important to assign a possible role for some of the genes of interest and to predict their cellular location and exposure to the immune system.
Extensive literature searches allow us to eliminate previously published genes by other groups. As result of this work we generated a list of conserved protein candidates, which might be located at the outer membrane of the meningococcus and/or display immunogenic properties. Additionally, functional tests were also conducted to demonstrate the ability of the antibodies generated against one of these antigens to induce complement-mediated bacterial death. The complement fixation assay is considered as the gold standard to assess the potential protective capacity of antibodies against N. meningitidis [19].
MATERIALS AND METHODS
Bacterial strains and growth conditions
For recombinant protein expression, W3110 Escherichia coli strain (New England BioLabs) was transformed with previously obtained genetic constructions to express each recombinant NMB1126 or NMB0181 proteins [20, 21], and were grown in M9 synthetic medium supplemented with 100 μg/mL ampicillin and additives as summarized in table 1.
N. meningitidis strains CU385 (B:4,7:P1.19,15:ST-33), NZ124 (B:4:P1.7b,4:ST-41) and C233 (C:2a:P1.9:ST-11) obtained from Finlay Institute in Havana, were used for Western blotting assay. One of them, the CU385 strain was also used for bactericidal test. In general, all neisserial strains were grown in a humidified atmosphere of 5% CO2 on brain heart infusion (BHI) agar (Oxoid, Basingstoke, United Kingdom) supplemented with an antibiotic mixture of vancomycin, colistin and nystatin (VCN, Oxoid) at the concentration recommended by the manufacturer.
Computer analysis
A number of computational algorithms free available in internet were used to search for signatures characteristic of putative OMPs and virulence related proteins from those genes annotated as ‘hypothetical’, ‘conserved hypothetical’ or with putative functions related to the cell envelope in the genome of MC58 N. meningitidis strain (Refseq NC_003112) (Figure 1). Genes with reported phase variation or homologous to human sequences were firstly excluded. When ORF products had more than one paralogs in the genome, only one of them was included. Homology searches were performed using the Blast program and non redundant databases. Conserved domains and sequences, as well as structural motifs, were identified by integrating COG, PFAM, PEDANT, PDB, SMART and CDD databases searches [22-24]. MBGD database was used to study the neighborhood of the genes of interest [25]. The degree of conservation of the selected genes was the most important criterion for the selection of a potential cross protective antigen. This parameter was analyzed from three N. meningitidis genomes: serogroup B strain MC58, serogroup A strain Z2491 (Refseq NC_003116.1) and serogroup C FAM18 (Refseq NC_008767). SignalP2.0 and LipoP programs [26, 27] were used to identify potential signal peptides. PSORT was used to predict cellular localization of the gene products [28]. Extensive literature search was carried out along all the process to avoid the selection of previously studied proteins by other researcher groups.
Expression and purification in E. coli
For expression of the nmb1126 and nmb0181 genes, previously obtained genetic constructions were used [20]. Recombinant proteins were expressed as fusion products with the N-terminal stabilizing segment of the P64k protein of N. meningitidis. They also contained a C-terminal His-tag fusion to facilitate the purification by IMAC (Immobilized metal ion affinity chromatography) according to manufacturer’s instructions (Amersham Pharmacia Biotech). Specific conditions to achieve each purification procedure are shown in table 1. A final step of dialysis against phosphate buffer, pH 8.0 was carried out in both cases.
Immunization schedule
Six weeks old BALB/c female mice, ten mice per group, were immunized with 20 μg of the NMB1126 or NMB0181 recombinant antigens. Preimmune sera were taken one day before the beginning of the schedule. The proteins were administered subcutaneously, in Freund’s Complete Adjuvant for the first dose and Freund’s Incomplete Adjuvant for the second (7 days) and third (14 days) doses. Animals were bled on day 21 for the corresponding analyses.
Western Blot against whole cell
To assess the recognition of the native proteins, 10 μg of whole cells from the CU385 strain, grown under the previously described conditions, were loaded into the SDS-PAGE gel and transferred to a nitrocellulose membrane (Hybond ECL; Amersham Pharmacia Biotech). Membranes were blocked with 5% skin milk in PBS and incubated with a 1:100 dilution of the antisera during 3h followed by a 1:5000 dilution of HPR-labeled anti–mouse IgG (Sigma). The revealing steps and analyses were performed as described previously [29].
Complement-mediated Bactericidal Activity
Serum bactericidal activity against CU385 N. meningitidis strain was evaluated as previously described [30]. Briefly, a pool of baby rabbit sera, previously screened for anti-meningococcal activity, was used as complement source. The bacterial suspension was adjusted to OD600nm 1.0 in phosphate buffered saline pH 8.0 (PBS) and incubated for 30 min at 56 ºC. A bacterial pellet corresponding to 2.5 mL of heat-inactivated bacterial suspension was added to 1.0 mL of rabbit serum and the mixture was incubated for 50 min on ice with agitation. Finally, bacteria were removed by centrifugation in a refrigerated centrifuge. If the antiserum shows a 50% reduction in the number of CFU per mL after incubation as compared to the preimmune serum at the same dilution, then is considered as positive.
RESULTS
In silico selection of putative vaccine candidates
The genome of N. meningitidis serogroup B strain MC58 was examined in a search for novel immune-exposed proteins. Genes encoding for proteins with a relative mass (Mr) outside the range between 8 kDa and 150 kDa were remove. ORF with unknown or hypothetical functions assigned (conserved or not), together with those primary annotated with a putative function related to the cell envelope were selected. A total of 725 sequences were retrieved.
Similitude to human proteins and phase variation in the expression of the genes were excluding criteria. In the case of duplicated ORF in the MC58 genome, one of the copies was removed to avoid redundancy. ORF products predicted by protein localization algorithms to encode for surface associated components were selected. Proteins homologous to those described as surface exposed in other bacteria were also chosen, independently of the in silico prediction.
ORF products resembling virulence-associated proteins were selected on the basis of sequence homology or the presence of motifs typical of such proteins. Other ORF products homologous to proteins previously reported as immunogenic, irrespective of their cellular location, were also retained. A degree of conservation higher than 90% in the Neisseria genomes studied was established as a requisite, aimed at the selection of candidates capable of inducing broadly reactive immunological responses.
Previously published proteins by other groups were identified by literature searching and manually removed.
ORFs encoding putative surface-exposed antigens identified as a result of the prediction of subcellular localization
Several approaches were used to determine the ORFs likely to encode proteins that are exposed at the bacterial surface, thus representing potential vaccine components. Of all sequences tested, 230 encoded for products with putative signal sequences for membrane proteins that are cleaved by signal peptidase I or signal peptidase II. PSORT server was next used to predict the cellular localization of these proteins. A total of 93 proteins were selected based on its potential localization at the outer membrane. They were examined in detail in terms of homology, degree of conservation and genomic localization. Finally, a set of 28 genes was selected and subdivided into unique Neisseria genes or orthologous ORF products (Table 2).
Putative new annotations were assigned to four genes based on overall homology and analysis of the conserved domains: nmb1333, that belongs to COG4942 (Membrane-bound metallopeptidase); nmb0506 included in DUF637 (possible hemagglutinin), nmb1964 related to gnl|CDD|8313, pfam02470 (mce related protein); and nmb0454 (tou4). In the cases of nmb1333 and nmb0506, possible neisserial pathogenic attributes, the assignment of functions was possible due to local homologies detected in some domains.
The nmb1333 gene appears to be the result of mutational events that eliminated a stop codon located upstream of the reported ATG initiation codon, resulting in the gain of more than 200aa at the N terminus. A similar event was detected for ORF nmb1470, with a corresponding addition of 70 aa., although no function could be assigned in this particular case. Additionally, the nmb0506 gene could have arisen from a fusion event between two adjacent genes as has been observed by neighborhood analyses for other genus (data not shown).
Some genes were found only in the serogroup B MC58 strain and could represent exclusive pathogenic attributes for this serogroup (genes nmb1008, nmb1010 and nmb1765).
Products selected for homologies to previously reported OMP or immunogenic orthologous proteins
In addition to the previous putative outer membrane proteins selected, we searched for gene products with homologies to previously reported OMPs, virulence attributes or immunogenic proteins relevant to vaccine design. According to this concept, four genes were characterized and finally selected (Table 3). A brief characterization of each gene follows.
nmb0181: Was annotated as a putative outer membrane protein H in the MC58. The homology was corroborated by its inclusion in the pfam03938 group “OmpH, Outer membrane protein (OmpH-like)” suggesting a common phylogenetically related ancestor for it and all the homologues. The outer membrane localization of OmpH proteins has been previously documented [31, 32]. Recently, these proteins have been associated with protection against haemorrhagic septicaemia after vaccination of calves with a live-attenuated aroA derivative of Pasteurella multocida B:2 [33].
nmb1483: Contains various conserved motifs related with cell wall biogenesis, including two LysM, a lysin domain found in a variety of enzymes involved in bacterial cell wall biogenesis, and a conserved region that belongs to the M23 peptidase family. Homologies to NlpD proteins have also been detected. This kind of proteins has been reported as an important virulence factor in Yersinia pestis [34].
nmb1126: Primary annotated as a hypothetical protein, it is now proposed to be re-annotated as the putative curli production assembly/transport component CsgG. Although neighborhood analyses showed no conserved gene organization as compared to other genus, its inclusion in COG1462 supports this conclusion. CsgG is an outer membrane lipoprotein, highly resistant to protease digestion and required to maintain the stability of CsgA and CsgB, structural components of the outer cell structure named curli.
nmb1693: Phylogenetically related to AsmA proteins (COG2982), an uncharacterized protein involved in outer membrane biogenesis. Recently, a role for Salmonella enterica AsmA protein in the invasion of epithelial cells have been suggested by Prieto et al. [35] (Table 4).
Immunization and immune response
Proteins NMB1126 and NMB0181 have been previously reported by our group as potential vaccines candidates following an alternative bioinformatic methodology based on the prediction of putative MHC class II epitopes [21]. Taking into account that both proteins have been selected by the two in silico approaches, we decided to proceed to experimental evaluation of their immunogenic properties. After successful expression in W3110 strain of E. coli, the proteins were purified by IMAC under denaturing conditions (Table 1).
To determine whether the purified recombinant proteins were capable of inducing a functional immune response, BALB/c mice were immunized as described before. The proteins were immunogenic in adult mice, eliciting IgG antibody titers around 1:70 000 and 1:60 000 for recombinant NMB1126 and NMB0181 respectively (data not shown).
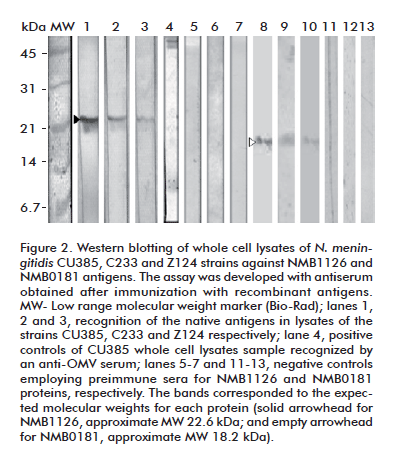
The sera from immunized animals were also capable of recognizing the native counterparts of the antigens in whole-cell lysates of three different N. meningitidis strains, including one of the serogroup C (Figure 2).
Bactericidal assays were next conducted to test the functional capabilities of the generated antibodies. Positive result was achieved for the recombinant antigens, with bactericidal titers of 1:128 for NMB1126 and 1:64 for NMB0181.
DISCUSSION
The discovery of a vaccine to prevent the disease caused by N. meningitidis serogroup B remains a priority in the fight against bacterial meningitis. Many strategies have been implemented so far in the search for an effective vaccine preparation, however, a definitive solution to this problem has not yet been found.
Amid this scenery, reverse vaccinology emerges as a time saving alternative to standard experimental searches for vaccine candidates [36]. The availability of the genomic sequence of the strain MC58 serogroup B from the year 2000 has facilitated the application of this methodology.
Pizza et al. searched the N. meningitidis genome looking for possible outer membrane proteins displaying overall sequence homology or sharing specific motifs with known antigens [9]. They combined these efforts with high throughput techniques to examine the immunogenic properties of more than 340 proteins. Those antigens with protective or bactericidal activity in vitro and/or in vivo were published. This strategy allowed this group of researchers to move towards the evaluation of a multicomponent vaccine, based on 5 of these proteins, which is currently in phase III clinical trial [10, 11]. Although in some cases the individual candidates selected showed up to 54% sequence variation, they combination was able to induce cross-reactive antibodies against a panel that covered the broad spectrum of bacterial strains circulating in the world [11, 37].
The reverse vaccinology approach, as has been implemented thus far, requires a large number of animals and is very labour and time intensive [17]. Unlike Pizza et al., we fixed a homology level above 90% among the sequenced genomes of N. meningitidis as one of the fundamental criteria for the selection of the candidate genes. This is a good a priori indicator for a high probability of generating a cross-reactive antibody response among different bacterial strains. The application of this criterion was expected to reduce the number of individual proteins needed for a multicomponent vaccine, with the consequent reduction of costs and complexity in its production process.
To initiate the prediction pipeline we only selected genes whose functions had not been fully elucidated and were annotated with hypothetical or putative functions. This selection gave us the advantage of working with less explored candidates.
The use of more than one software application to predict possible signal peptide sequences contributed to generate more reliable information. The LipoP server also enabled a safest prediction of lipoproteins, which have been widely documented as important vaccine candidates (e.g., OspA of Borrelia burgdorferi P6 of Haemophilus influenzae and GNA1870 of N. meningitidis) [13, 38-40]. Lipoproteins located at the outer membrane also offer advantages in setting up purification procedures as compared to the typical integral beta barrel proteins of Gram-negative bacteria [41].
The study of the genomic neighborhood, in conjunction with the analyses of conserved domains and the homology through different servers is also a powerful tool for the assignment and annotation of genomes [42]. Proteins displaying some degree of homology or belonging to the same orthologous cluster, and that additionally have their gene neighborhood preserved, have a high potential to perform similar functions. In this sense, the structural alignment with homologous proteins of known function could provide additional elements to support the initial results.
The inclusion of ORF products without homology with other known proteins implies the risk of being false ORFs, but also maximizes the possibility of selecting genus specific pathogenic attributes, which is a desirable goal. Some of the main vaccine candidates proposed for the prevention of meningococcal disease have included proteins with similar characteristics. One of them, the GNA1870, is one of the most studied and successful so far [13].
In this article we presented the results of the immunological evaluation of two proteins that were identified both by a previously MHC class II epitopes prediction and by the in silico methodology proposed hear. Previously, in the article of Pajón et al., both proteins have been cloned, purified, and used to immunize mice [21]. Nevertheless, the details of each mentioned process and the results were only drawback. In this article we report the exact corresponding data. These proteins were also identified by several proteomic techniques in the study of OMV from neisserial species. At these respect, both proteins were detected in the composition of OMV from a gna33 N. meningitidis serogroup B mutant. The authors show that these OMV are able to induce bactericidal antibodies with broader cross-protective activities than the OMV obtained from the non mutant corresponding strain [43]. Also NMB1126 protein was detected in New Zealand OMV vaccine from strain NZ98/254 and in OMV from a N. lactamica, another alternative study to achieve protection against the meningococcus [44, 45]. More recently, Tsolakos et al. founded that in some growth conditions, various OMP included NMB1126 increased significantly its representativity in bacterial OMV, a fact that was associated with higher serum bactericidal titres in mice immunized with corresponding OMV vaccine [46]. According to our results, both proteins could be contributing to this improvement in immune responses.
Despite the evident importance and benefits of using in silico predicting tools, any outcome from these methods would require experimental corroboration before the final endorsement. A successful vaccine must be able to induce a functional response in mice, measured in either in vitro and/or in vivo tests. Ideally, those tests must be good correlates of protection in humans [47]. The assay of complement mediated bactericidal activity is a relatively rapid in vitro test, considered as the gold standard for N. meningitidis [48]. The fact that the candidate NMB1126 was capable of induces titres of 1:128 against the homologous strain is a good indication that supports their subsequent evaluation. Also, the recognition of the native antigen in different strains of the same and different serogroups is a good indicator of the possibility of induces functional response against diverse meningococci, a criterion that is considered essential in the anti-meningococcal vaccine field. This antigen could be included in a vaccine preparation, although a deeper characterization is needed.
The results shown above are a good example of how in silico and experimental analyses can be combined to identify novel antigens whose existence has just been proposed. The proposed methodology includes new selection strategies as compared with previous works. The proposed candidates have the potentiality to integrate a universal vaccine against the meningococcal disease.
The ability of bioinformatics to characterize genomic sequences from pathogenic bacteria for the prediction of genes that may encode vaccine candidates (e.g., surface localized proteins) has been corroborated. Our study demonstrated that bioinformatics is a very useful tool to expedite the vaccine discovery process in N. meningitidis by rapidly providing a set of uncharacterized candidates for further testing.
ACKNOWLEDGMENTS
The authors are very grateful to Dr. Gerardo Guillén, Dr. Carlos Duarte and Dr. Ricardo Bringas for their critical reading and useful comments in the revision of the manuscript.
REFERENCES
1. Stephens DS, Zimmer SM. Pathogenesis, Therapy, and Prevention of Meningococcal Sepsis. Curr Infect Dis Rep. 2002;4(5):377-86.
2. Tikhomirov E, Santamaria M, Esteves K. Meningococcal disease: public health burden and control. World Health Stat Q. 1997;50(3-4):170-7.
3. Finne J, Leinonen M, Makela PH. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet. 1983;2(8346):355-7.
4. Fredriksen JH, Rosenqvist E, Wedege E, Bryn K, Bjune G, Froholm LO, et al. Production, characterization and control of MenB-vaccine "Folkehelsa": an outer membrane vesicle vaccine against group B meningococcal disease. NIPH Ann. 1991;14(2):67-80.
5. Sierra GV, Campa HC, Varcacel NM, Garcia IL, Izquierdo PL, Sotolongo PF, et al. Vaccine against group B Neisseria meningitidis: protection trial and mass vaccination results in Cuba. NIPH Ann 1991;14(2):195-10.
6. Perkins BA, Jonsdottir K, Briem H, Griffiths E, Plikaytis BD, Hoiby EA, et al. Immunogenicity of two efficacious outer membrane protein-based serogroup B meningococcal vaccines among young adults in Iceland. J Infect Dis. 1998;177(3):683-91.
7. Wheeler JX, Vipond C, Feavers IM. Exploring the proteome of meningococcal outer membrane vesicle vaccines. Proteomics Clin Appl. 2007;1(9):1198-210.
8. Masignani V, Rappuoli R, Pizza M. Reverse vaccinology: a genome-based approach for vaccine development. Expert Opin Biol Ther. 2002;2(8):895-905.
9. Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287(5459):1816-20.
10. Snape MD, Dawson T, Morant A, John B, Ohene-Kena R, Borrow R, et al. Program and abstract of the 16th International Pathogenic Neisseria Conference (Rotterdam, Amsterdam). 7-12 September. 2008. Immunogenicity and reactogenicity of a novel serogroup B Neisseria meningitidis vaccine administered from 6 months of age. Abstract O69.
11. Giuliani MM, Adu-Bobie J, Comanducci M, Arico B, Savino S, Santini L, et al. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci USA. 2006;103(29):10834-9.
12. Capecchi B, Adu-Bobie J, Di Marcello F, Ciucchi L, Masignani V, Taddei A, et al. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol Microbiol. 2005;55(3):687-98.
13. Masignani V, Comanducci M, Giuliani MM, Bambini S, Adu-Bobie J, Arico B, et al. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med. 2003;197(6):789-99.
14. Ruckdeschel EA, Kirkham C, Lesse AJ, Hu Z, Murphy TF. Mining the Moraxella catarrhalis genome: identification of potential vaccine antigens expressed during human infection. Infect Immun. 2008;76(4):1599-607.
15. Huntley JF, Conley PG, Hagman KE, Norgard MV. Characterization of Francisella tularensis outer membrane proteins. J Bacteriol. 2007;189(2):561-74.
16. Lewenza S, Gardy JL, Brinkman FS, Hancock RE. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 2005;15(2):321-9.
17. Zhao Y, Benita Y, Lok M, Kuipers B, van der Ley P, Jiskoot W, et al. Multi-antigen immunization using IgG binding domain ZZ as carrier. Vaccine. 2005;23(43):5082-90.
18. Pajon R, Yero D, Niebla O, Climent Y, Sardinas G, Garcia D, et al. Identification of new meningococcal serogroup B surface antigens through a systematic analysis of neisserial genomes. Vaccine. 2009;28(2):532-41.
19. Borrow R, Carlone GM, Rosenstein N, Blake M, Feavers I, Martin D, et al. Neisseria meningitidis group B correlates of protection and assay standardization--international meeting report Emory University, Atlanta, Georgia, United States, 16-17 March 2005. Vaccine. 2006;24(24):5093-107.
20. Yero D, Pajon R, Niebla O, Sardinas G, Vivar I, Perera Y, et al. Bicistronic expression plasmid for the rapid production of recombinant fused proteins in Escherichia coli. Biotechnol Appl Biochem. 2006;44(Pt 1):27-34.
21. Pajon R, Yero D, Niebla O, Climent Y, Sardinas G, Garcia D, et al. Identification of new meningococcal serogroup B surface antigens through a systematic analysis of neisserial genomes. Vaccine. 2009;28(2):532-41.
22. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389-402.
23. Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30(1):281-3.
24. Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res 2007;35(Database issue):D237-40.
25. Uchiyama I. MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucleic Acids Res. 2007;35(Database issue):D343-6.
26. Nielsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc Int Conf Intell Syst Mol Biol. 1998;6:122-30.
27. Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12(8):1652-62.
28. Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24(1):34-6.
29. Wedege E, Froholm LO. Human antibody response to a group B serotype 2a meningococcal vaccine determined by immunoblotting. Infect Immun. 1986;51(2):571-8.
30. Delgado M, Yero D, Niebla O, Gonzalez S, Climent Y, Perez Y, et al. Lipoprotein NMB0928 from Neisseria meningitidis serogroup B as a novel vaccine candidate. Vaccine. 2007;25(50):8420-31.
31. Luo Y, Glisson JR, Jackwood MW, Hancock RE, Bains M, Cheng IH, et al. Cloning and characterization of the major outer membrane protein gene (ompH) of Pasteurella multocida X-73. J Bacteriol. 1997;179(24):7856-64.
32. Koski P, Hirvas L, Vaara M. Complete sequence of the ompH gene encoding the 16-kDa cationic outer membrane protein of Salmonella typhimurium. Gene. 1990;88(1):117-20.
33. Ataei S, Burchmore R, Christopher Hodgson J, Finucane A, Parton R, Coote JG. Identification of immunogenic proteins associated with protection against haemorrhagic septicaemia after vaccination of calves with a live-attenuated aroA derivative of Pasteurella multocida B:2. Res Vet Sci. 2009;87(2):207-10.
34. Tidhar A, Flashner Y, Cohen S, Levi Y, Zauberman A, Gur D, et al. The NlpD lipoprotein is a novel Yersinia pestis virulence factor essential for the development of plague. PLoS One. 2009;4(9):e7023.
35. Prieto AI, Hernandez SB, Cota I, Pucciarelli MG, Orlov Y, Ramos-Morales F, et al. Roles of the outer membrane protein AsmA of Salmonella enterica in the control of marRAB expression and invasion of epithelial cells. J Bacteriol. 2009;191(11):3615-22.
36. Adu-Bobie J, Capecchi B, Serruto D, Rappuoli R, Pizza M. Two years into reverse vaccinology. Vaccine. 2003;21(7-8):605-10.
37. Jacobsson S, Thulin S, Molling P, Unemo M, Comanducci M, Rappuoli R, et al. Sequence constancies and variations in genes encoding three new meningococcal vaccine candidate antigens. Vaccine. 2006;24(12):2161-8.
38. Murphy TF, Kirkham C, Lesse AJ. Construction of a mutant and characterization of the role of the vaccine antigen P6 in outer membrane integrity of nontypeable Haemophilus influenzae. Infect Immun. 2006;74(9):5169-76.
39. Sigal LH, Zahradnik JM, Lavin P, Patella SJ, Bryant G, Haselby R, et al. A vaccine consisting of recombinant Borrelia burgdorferi outer-surface protein A to prevent Lyme disease. Recombinant Outer-Surface Protein A Lyme Disease Vaccine Study Consortium. N Engl J Med. 1998;339(4):216-22.
40. Steere AC, Sikand VK, Meurice F, Parenti DL, Fikrig E, Schoen RT, et al. Vaccination against Lyme disease with recombinant Borrelia burgdorferi outer-surface lipoprotein A with adjuvant. Lyme Disease Vaccine Study Group. N Engl J Med. 1998;339(4):209-15.
41. Booth PJ. The trials and tribulations of membrane protein folding in vitro. Biochim Biophys Acta. 2003;1610(1):51-6.
42. Fong C, Rohmer L, Radey M, Wasnick M, Brittnacher MJ. PSAT: a web tool to compare genomic neighborhoods of multiple prokaryotic genomes. BMC Bioinformatics. 2008;9:170.
43. Ferrari G, Garaguso I, Adu-Bobie J, Doro F, Taddei AR, Biolchi A, et al. Outer membrane vesicles from group B Neisseria meningitidis delta gna33 mutant: proteomic and immunological comparison with detergent-derived outer membrane vesicles. Proteomics. 2006;6(6):1856-66.
44. Vipond C, Suker J, Jones C, Tang C, Feavers IM, Wheeler JX. Proteomic analysis of a meningococcal outer membrane vesicle vaccine prepared from the group B strain NZ98/254. Proteomics. 2006;6(11):3400-13.
45. Vaughan TE, Skipp PJ, O'Connor CD, Hudson MJ, Vipond R, Elmore MJ, et al. Proteomic analysis of Neisseria lactamica and N eisseria meningitidis outer membrane vesicle vaccine antigens. Vaccine. 2006;24(25):5277-93.
46. Tsolakos N, Lie K, Bolstad K, Maslen S, Kristiansen PA, Hoiby EA, et al. Characterization of meningococcal serogroup B outer membrane vesicle vaccines from strain 44/76 after growth in different media. Vaccine. 2010;28(18):3211-8.
47. Moxon R, Rappuoli R. Bacterial pathogen genomics and vaccines. Br Med Bull. 2002;62:45-58.
48. Goldschneider I, Gotschlich EC, Artenstein MS. Human immunity to the meningococcus. I. The role of humoral antibodies. J Exp Med. 1969;129(6):1307-26.
49. Vasfi Marandi M, Harel J, Mittal KR. Identification by monoclonal antibodies of serotype D strains of Pasteurella multocida representing various geographic origins and host species. J Med Microbiol. 1997;46(7):603-10.
50. Vemulapalli R, Duncan AJ, Boyle SM, Sriranganathan N, Toth TE, Schurig GG. Cloning and sequencing of yajC and secD homologs of Brucella abortus and demonstration of immune responses to YajC in mice vaccinated with B. abortus RB51. Infect Immun. 1998;66(12):5684-91.
Received in December, 2010.
Accepted for publication in February, 2012.
Darien García. Meningococcal Research Department, Division of Vaccines, Center for Genetic Engineering and Biotechnology, CIGB. Ave. 31, Cubanacan, PO Box 10 600, Havana, Cuba. E-mail: darien.garcia@cigb.edu.cu.